4 Data Wrangling
4.1 R setup for this chapter
This section loads all needed R packages for this chapter. Appendix A lists all R packages used in this book, and also provides R session information. Appendix B describes the 431-Love.R script, and demonstrates its use.
4.2 Data from a .csv file: Cleveland Neighborhoods
Appendix C provides further guidance on pulling data from other systems into R, while Appendix D gives more information (including download links) for all data sets used in this book.
We will demonstrate ideas in this chapter using a tibble containing information on neighborhoods within the city of Cleveland, Ohio. Here, we ingest the data from a .csv (comma-separated version) file called cle_nbd.csv into an R tibble we’ll call cle_neigh, then print the result.
cle_neigh <- read_csv("data/cle_nbd.csv",
show_col_types = FALSE
)
cle_neigh# A tibble: 34 × 9
nbhd_id nbhd_name pop20 pop10 pop_grow location age65plus income22 unemp22
<dbl> <chr> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr>
1 1 BELLAIRE-PU… 13823 13380 Rise West 13.8 41324 Middle
2 2 BROADWAY-SL… 18854 22331 Fall Central 11.8 32896 Middle
3 3 BROOKLYN CE… 8315 8948 Fall Central 11.8 32308 Middle
4 4 BUCKEYE SHA… 11419 12470 Fall East 22.5 35402 Middle
5 5 CENTRAL 11955 12306 Fall Central 7.41 16258 High
6 6 CLARK FULTON 7625 8509 Fall Central 10.6 33006 Middle
7 7 COLLINWOOD … 9616 11542 Fall East 13.5 30580 Middle
8 8 CUDELL 9125 9287 Fall West 10.3 29417 High
9 9 CUYAHOGA VA… 1404 1371 Rise Central 1.54 20266 High
10 10 DETROIT-SHO… 11285 11577 Fall Central 12.0 45235 Low
# ℹ 24 more rowsn_miss(cle_neigh)[1] 0Key sources for these data were U.S. Census Bureau (2020) and NEOCANDO (2024). The data in the cle_neigh tibble describe each of the 34 neighborhoods (technically statistical planning areas) of the City of Cleveland. For each neighborhood, the cle_neigh tibble contains 9 variables, described in the table below.
| Variable | Description |
|---|---|
| nbhd_id | alphabetical order (1-34) of Cleveland neighborhoods |
| nbhd_name | name of neighborhood |
| pop20 | Total Population, from Decennial Census 2020 |
| pop10 | Total Population, from Decennial Census 2010 |
| pop_grow | Rise if Total population increased from 2010 to 2020, else Fall |
| location | neighborhood’s geographic location (East, Central or West) |
| age65plus | % of residents ages 65 and higher, from ACS |
| income22 | Median household income, from ACS |
| unemp22 | Unemployment rate as Low (< 10%), Middle (10-15%), High (above 15%), from ACS |
- ACS refers to 5-year estimates from the American Community Survey, covering 2018-2022.
4.3 Key Functions for Managing Data
The dplyr package, part of the tidyverse, provides a set of functions (verbs) which we will use frequently in this course to manage data.
4.3.1 The pipe
There are two main pipes used in R. They are tools for clearer coding that is easier to read, when you are putting multiple steps together into chain.
For example, the code below uses the |> pipe:
cle_neigh |> head()# A tibble: 6 × 9
nbhd_id nbhd_name pop20 pop10 pop_grow location age65plus income22 unemp22
<dbl> <chr> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr>
1 1 BELLAIRE-PUR… 13823 13380 Rise West 13.8 41324 Middle
2 2 BROADWAY-SLA… 18854 22331 Fall Central 11.8 32896 Middle
3 3 BROOKLYN CEN… 8315 8948 Fall Central 11.8 32308 Middle
4 4 BUCKEYE SHAK… 11419 12470 Fall East 22.5 35402 Middle
5 5 CENTRAL 11955 12306 Fall Central 7.41 16258 High
6 6 CLARK FULTON 7625 8509 Fall Central 10.6 33006 Middle applies the head() function to the cle_neigh data. Specifically, the |> pipes the information from the cle_neigh data into the function head(). As we’ll see, we can string a series of pipes together to apply a chain of actions to our data.
The head() function simply shows the first 6 rows of the data, while the tail() function shows the last 6 rows.
Another, similar, pipe is %>%, which does almost the same thing as |> and is read the same way. Here, we pipe the cle_neigh data into the tail() function, and also ask to show the last 8 rows of the data, instead of the default six.
# A tibble: 8 × 9
nbhd_id nbhd_name pop20 pop10 pop_grow location age65plus income22 unemp22
<dbl> <chr> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr>
1 27 OLD BROOKLYN 32315 32009 Rise Central 13.3 48061 Middle
2 28 SAINT CLAIR-… 5139 6876 Fall East 16.7 29612 Middle
3 29 STOCKYARDS 9522 10411 Fall Central 12.3 33553 Middle
4 30 TREMONT 7699 7975 Fall Central 9.44 58716 Low
5 31 UNION-MILES … 15625 19004 Fall East 20.1 32788 High
6 32 UNIVERSITY C… 9558 7939 Rise East 14.9 24780 Low
7 33 WEST BOULEVA… 18971 18888 Rise West 9.75 37973 Middle
8 34 WOODLAND HIL… 5625 6678 Fall East 19.0 25696 Middle Wickham, Çetinkaya-Rundel, and Grolemund (2024) has much more on the pipe in its Pipes section.
4.3.2 select()
We use the select() function to choose specific variables (columns) from our tibble.
cle_neigh |>
select(nbhd_name, pop_grow)# A tibble: 34 × 2
nbhd_name pop_grow
<chr> <chr>
1 BELLAIRE-PURITAS Rise
2 BROADWAY-SLAVIC VILLAGE Fall
3 BROOKLYN CENTRE Fall
4 BUCKEYE SHAKER Fall
5 CENTRAL Fall
6 CLARK FULTON Fall
7 COLLINWOOD NOTTINGHAM Fall
8 CUDELL Fall
9 CUYAHOGA VALLEY Rise
10 DETROIT-SHOREWAY Fall
# ℹ 24 more rowsAs you can see, this version of the data includes only the two variables we selected.
4.3.3 filter()
In contrast, we use the filter() function to choose specific subjects (rows) from our tibble.
cle_neigh |>
filter(pop_grow == "Rise" & location == "West")# A tibble: 5 × 9
nbhd_id nbhd_name pop20 pop10 pop_grow location age65plus income22 unemp22
<dbl> <chr> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr>
1 1 BELLAIRE-PUR… 13823 13380 Rise West 13.8 41324 Middle
2 12 EDGEWATER 6000 5851 Rise West 13.5 52709 Low
3 17 HOPKINS 366 215 Rise West 14.5 29497 Middle
4 19 JEFFERSON 17351 16548 Rise West 11.3 49908 Low
5 33 WEST BOULEVA… 18971 18888 Rise West 9.75 37973 Middle Here, we’ve selected only those rows that have rising population growth and are located in the West.
We can combine filter() and select() to choose both specific rows (here, those with incomes over 50,000) and columns (neighborhood ID code, name, income, unemployment and location.)
# A tibble: 6 × 5
nbhd_id nbhd_name income22 unemp22 location
<dbl> <chr> <dbl> <chr> <chr>
1 11 DOWNTOWN 61467 Low Central
2 12 EDGEWATER 52709 Low West
3 20 KAMMS CORNERS 63280 Low West
4 22 LEE-HARVARD 53278 Middle East
5 26 OHIO CITY 62890 Low Central
6 30 TREMONT 58716 Low Central 4.3.4 mutate()
We use the mutate() function to create new variables that are combinations of the old ones. For instance, here we create a pop_chg variable which is the percentage change in population from pop10 to pop20.
cle_neigh |>
mutate(pop_chg = 100 * (pop20 - pop10) / pop20) |>
select(nbhd_name, pop20, pop10, pop_grow, pop_chg) |>
tail(4)# A tibble: 4 × 5
nbhd_name pop20 pop10 pop_grow pop_chg
<chr> <dbl> <dbl> <chr> <dbl>
1 UNION-MILES PARK 15625 19004 Fall -21.6
2 UNIVERSITY CIRCLE 9558 7939 Rise 16.9
3 WEST BOULEVARD 18971 18888 Rise 0.438
4 WOODLAND HILLS 5625 6678 Fall -18.7 Here, we use the tail(4) function to show the last four rows in this new set of data.
Another common use of mutate() is to create transformations of our data, using functions like the square root, inverse or logarithm.
cle_neigh |>
mutate(loginc = log(income22)) |>
ggplot(aes(x = income22, y = loginc, col = location)) +
geom_point(size = 3) +
theme_bw()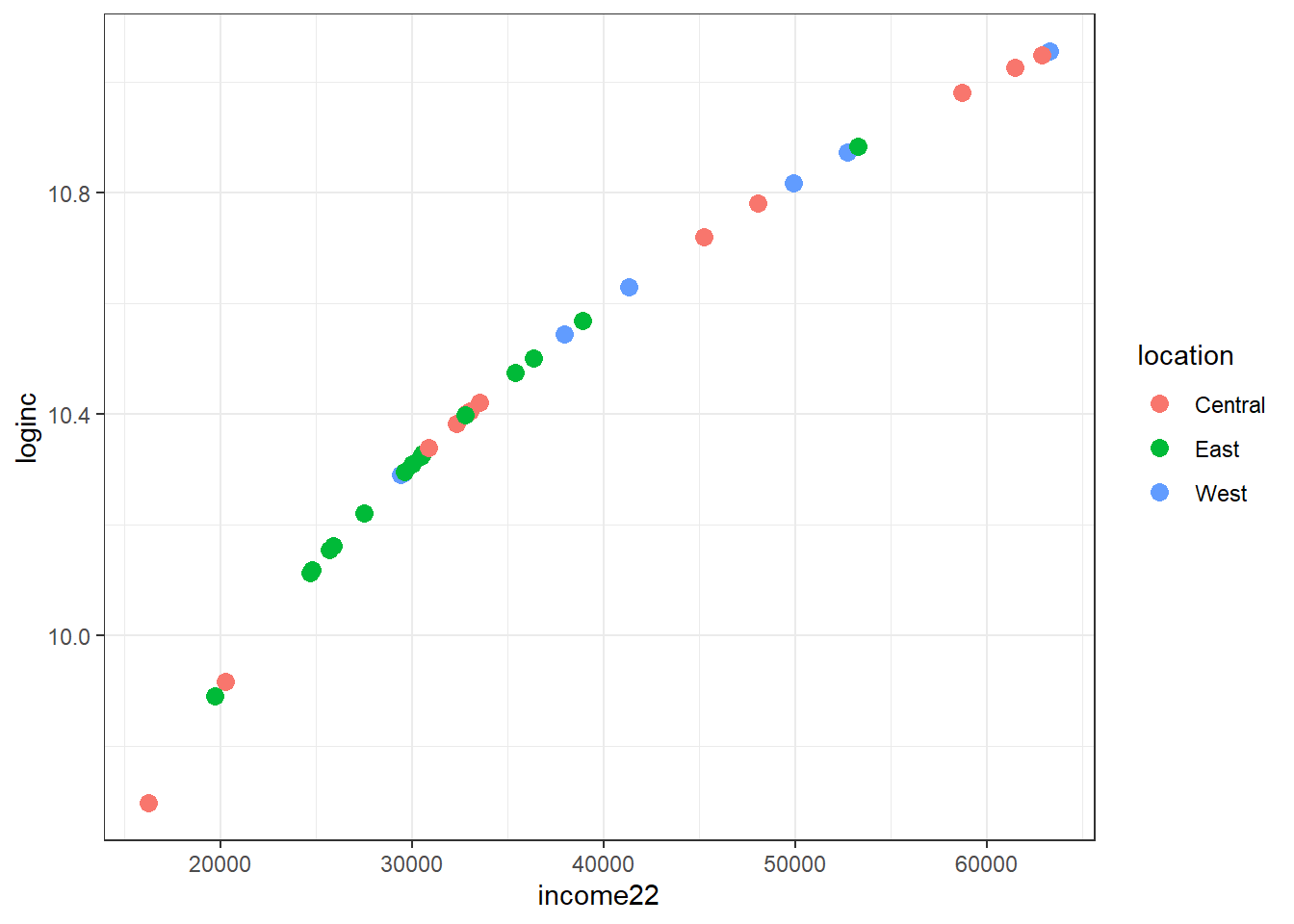
Note that log() is the natural logarithm in R, which we prefer to log10() (the base 10 logarithm) for most of our work because coefficients on the natural log scale will be more easily interpreted, as we’ll see later.
4.3.5 Creating Factors with mutate()
Sometimes we will want to change all of the character variables in a data set into factor variables in R. Factors are used for categorical variables, variables that have a fixed and known set of possible values. They are also useful when you want to display character vectors in a non-alphabetical order. To do so, my favorite approach is:
# A tibble: 34 × 9
nbhd_id nbhd_name pop20 pop10 pop_grow location age65plus income22 unemp22
<dbl> <fct> <dbl> <dbl> <fct> <fct> <dbl> <dbl> <fct>
1 1 BELLAIRE-PU… 13823 13380 Rise West 13.8 41324 Middle
2 2 BROADWAY-SL… 18854 22331 Fall Central 11.8 32896 Middle
3 3 BROOKLYN CE… 8315 8948 Fall Central 11.8 32308 Middle
4 4 BUCKEYE SHA… 11419 12470 Fall East 22.5 35402 Middle
5 5 CENTRAL 11955 12306 Fall Central 7.41 16258 High
6 6 CLARK FULTON 7625 8509 Fall Central 10.6 33006 Middle
7 7 COLLINWOOD … 9616 11542 Fall East 13.5 30580 Middle
8 8 CUDELL 9125 9287 Fall West 10.3 29417 High
9 9 CUYAHOGA VA… 1404 1371 Rise Central 1.54 20266 High
10 10 DETROIT-SHO… 11285 11577 Fall Central 12.0 45235 Low
# ℹ 24 more rowsNote that the nbhd_name, pop_grow and unemp22 variables are all now factors in R. We’ll discuss factors in more detail as the semester moves along.
4.3.6 arrange()
cle_neigh |>
mutate(pop_chg = 100 * (pop20 - pop10) / pop20) |>
select(nbhd_name, pop20, pop10, pop_grow, pop_chg) |>
arrange(pop_chg) |>
head()# A tibble: 6 × 5
nbhd_name pop20 pop10 pop_grow pop_chg
<chr> <dbl> <dbl> <chr> <dbl>
1 SAINT CLAIR-SUPERIOR 5139 6876 Fall -33.8
2 GLENVILLE 21137 27394 Fall -29.6
3 MOUNT PLEASANT 14015 17320 Fall -23.6
4 UNION-MILES PARK 15625 19004 Fall -21.6
5 FAIRFAX 5167 6239 Fall -20.7
6 COLLINWOOD NOTTINGHAM 9616 11542 Fall -20.0By default, the arrangement here is sorted from lowest (here, most negative) to highest. To sort from highest to lowest, we could substitute
arrange(desc(pop_chg))into the code above.
4.3.7 summarise() and group_by()
cle_neigh |>
group_by(location) |>
summarise(
n = n(), mean = mean(income22), sd = sd(income22),
med = median(income22), mad = mad(income22)
) |>
kable(digits = 2)| location | n | mean | sd | med | mad |
|---|---|---|---|---|---|
| Central | 12 | 39628.58 | 15556.57 | 33279.5 | 18509.52 |
| East | 15 | 31038.13 | 7954.50 | 30023.0 | 6415.21 |
| West | 7 | 43444.00 | 12561.80 | 41324.0 | 16879.40 |
4.4 Describing the Data
Consider the differences between the population in 2020 and the population in 2010 across these neighborhoods.
cle_neigh |>
reframe(lovedist(pop20 - pop10))# A tibble: 1 × 10
n miss mean sd med mad min q25 q75 max
<int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 34 0 -719. 1684. -519 819. -6257 -1052. 70.5 38384.4.1 Are population changes associated with location?
cle_neigh |>
reframe(lovedist(pop20 - pop10), .by = location)# A tibble: 3 × 11
location n miss mean sd med mad min q25 q75 max
<chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 West 7 0 101. 487. 149 436. -758 -39.5 297 803
2 Central 12 0 -171. 1641. -288. 696. -3477 -696. 101. 3838
3 East 15 0 -1540. 1774. -1053 1014. -6257 -1857 -698 16194.4.2 Re-ordering the levels of a factor
cle_neigh |>
tabyl(unemp22) unemp22 n percent
High 9 0.2647059
Low 11 0.3235294
Middle 14 0.4117647That’s not so helpful. We really want to show these categories in a natural order (either High then Middle then Low, or the opposite.) We can use the forcats package and the fct_relevel() function to help.
cle_neigh <- cle_neigh |>
mutate(
unemp22 =
fct_relevel(unemp22, "Low", "Middle", "High")
)
cle_neigh |>
tabyl(unemp22) unemp22 n percent
Low 11 0.3235294
Middle 14 0.4117647
High 9 0.26470594.4.3 Plotting three groups
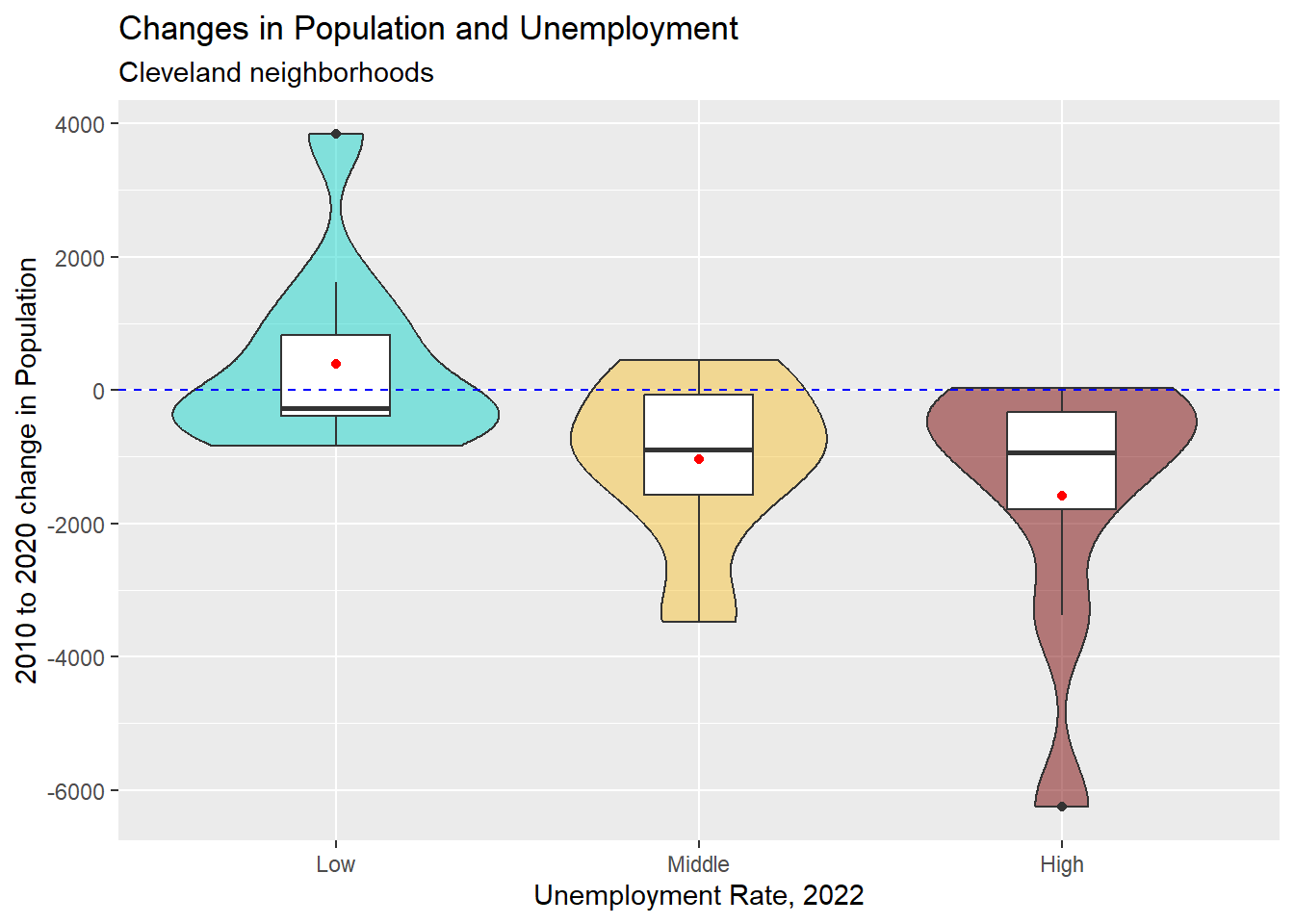
cle_neigh <- cle_neigh |>
mutate(pop_diff = pop20 - pop10)
ggplot(data = cle_neigh, aes(
x = unemp22, y = pop_diff,
fill = unemp22
)) +
geom_violin() +
geom_boxplot(fill = "white", width = 0.3) +
stat_summary(fun = "mean", geom = "point", col = "red") +
geom_hline(
yintercept = 0, col = "blue",
linetype = "dashed"
) +
scale_fill_viridis_d(
option = "turbo",
begin = 0.3, alpha = 0.5
) +
guides(fill = "none") +
labs(
title = "Changes in Population and Unemployment",
y = "2010 to 2020 change in Population",
x = "Unemployment Rate, 2022",
subtitle = "Cleveland neighborhoods"
)
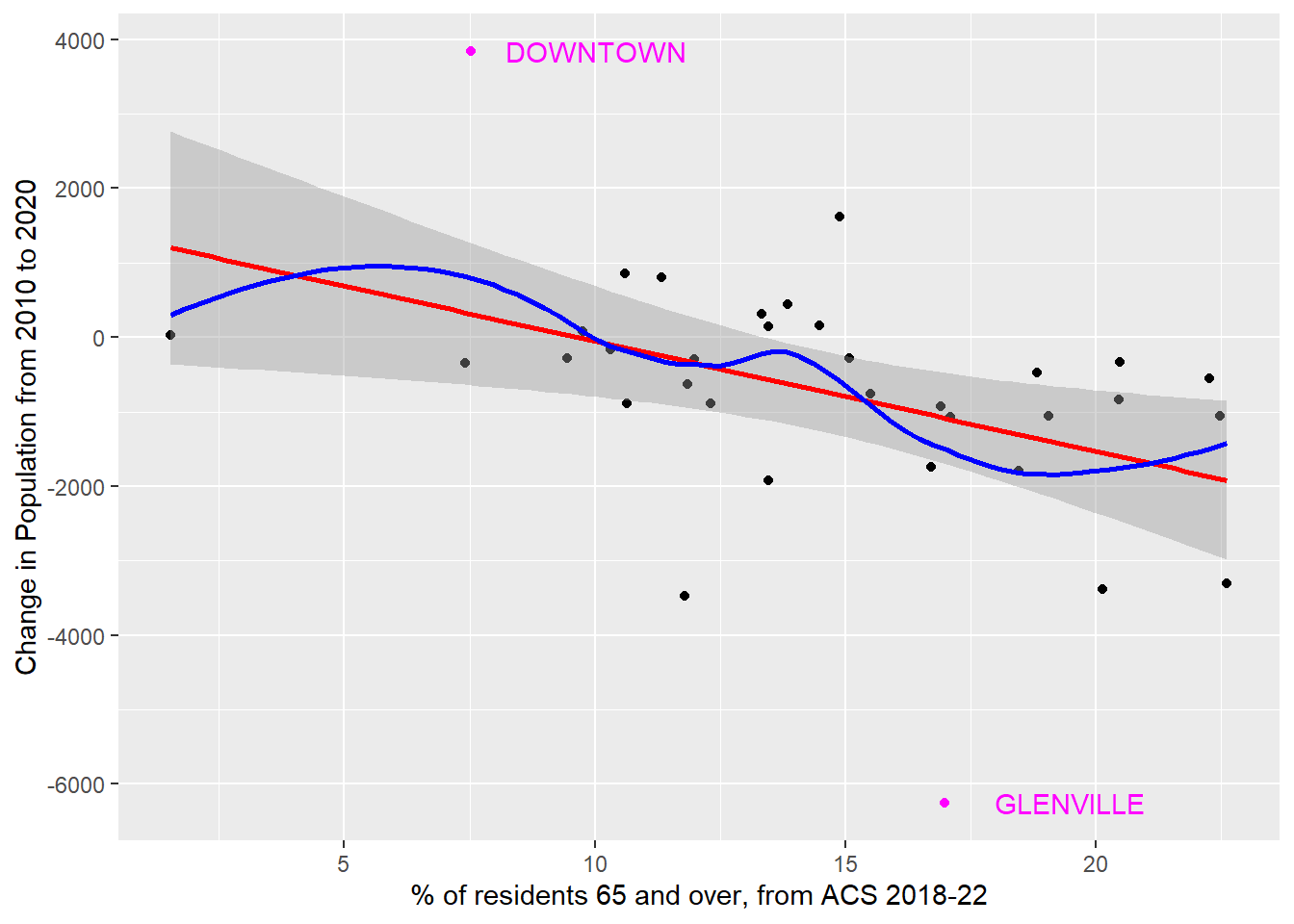
4.4.4 Association of Population Change and % 65+
ggplot(cle_neigh, aes(x = age65plus, y = pop_diff)) +
geom_point() +
geom_smooth(
method = "lm", formula = y ~ x,
se = TRUE, col = "red"
) +
geom_smooth(
method = "loess", formula = y ~ x,
se = FALSE, col = "blue"
) +
geom_text(
data = cle_neigh |>
filter(pop_diff > 3000 | pop_diff < -6000),
aes(label = nbhd_name),
nudge_x = 2.5, col = "magenta"
) +
geom_point(
data = cle_neigh |>
filter(pop_diff > 3000 | pop_diff < -6000),
col = "magenta"
) +
labs(
y = "Change in Population from 2010 to 2020",
x = "% of residents 65 and over, from ACS 2018-22"
)
4.5 Working with Factors
The forcats package is an important piece of the tidyverse, containing many functions which solve common problems with factors. This PDF cheat sheet is available to help you decide which of the forcats functions might be most useful to help you solve your current problem
Examples of forcats functions that we will use in this course include:
-
fct_recode(), which lets us change factor levels by hand -
fct_relevel(), which lets us reorder factor levels by hand -
fct_infreq(), which lets us reorder factor levels by their frequency -
fct_collapse(), which lets us collapse factor levels into manually defined groups -
fct_lump(), which lets us lump uncommon factor levels together into an “other” level
The best place to learn more about factors is the chapter on factors in Wickham, Çetinkaya-Rundel, and Grolemund (2024).
The best place to start learning more about forcats functions is this Introduction to forcats.
4.6 For More Information
The dplyr page is a comprehensive source of descriptions for the tools in the
dplyrpackage.The R Graphics Cookbook has an excellent chapter on Getting Your Data into Shape which is well worth your time.
R for Data Science (see Wickham, Çetinkaya-Rundel, and Grolemund (2024)) provides an excellent chapter on Data Transformation which goes into further detail on many of the issues discussed here. Other key chapters with something to say about these issues include:
- Workflow: basics
- Code Style and also Data tidying at https://r4ds.hadley.nz/data-tidy
- Factors
- The
datawizardpackage within the easystats eco-system (see Lüdecke et al. (2022)) has many useful functions.
- In particular, the Coming from Tidyverse page provides a nice vignette describing many of the easystats approaches for wrangling data.
- The
stylerpackage helps to format code according to the tidyverse style guide, which has some appealing features. I try to use this style throughout this book, to improve readability of the work, and I do so by applying a special add-in within R Studio.