22 Multiple Regression and Imputation
This is a sketchy draft with some useful content. I’ll remove this notice when I post a version of this Chapter that is essentially finished, but that won’t be until summer 2025.
22.1 R setup for this chapter
Appendix A lists all R packages used in this book, and also provides R session information. Appendix B describes the 431-Love.R script, and demonstrates its use.
22.2 Data will be Countries of the World version 2
Appendix C provides further guidance on pulling data from other systems into R, while Appendix D gives more information (including download links) for all data sets used in this book.
ctry <- read_csv("data/countries.csv", show_col_types = FALSE) |>
mutate(
UNIV_CARE = factor(UNIV_CARE),
WHO_REGION = factor(WHO_REGION),
WBANK_INCOME = factor(WBANK_INCOME),
C_NUM = as.character(C_NUM)
) |>
janitor::clean_names()
ctry# A tibble: 192 × 14
c_num country_name hale_all univ_care wbank_income hale_2000 births_attend
<chr> <chr> <dbl> <fct> <fct> <dbl> <dbl>
1 1 Afghanistan 50.4 no Low 46.8 68
2 2 Albania 66.7 yes Upper-middle 65.2 100
3 3 Algeria 65.5 yes Lower-middle 62.7 99
4 4 Andorra NA no High NA 100
5 5 Angola 53.8 no Lower-middle 42.9 50
6 6 Antigua and Ba… 66.5 no High 65.5 99
7 7 Argentina 64.8 yes Upper-middle 65.1 99
8 8 Armenia 64 no Upper-middle 63.5 100
9 9 Australia 70.6 yes High 68.6 99
10 10 Austria 69.8 yes High 68.2 98
# ℹ 182 more rows
# ℹ 7 more variables: ihr_core_cap <dbl>, avg_tempc <dbl>, female_22 <dbl>,
# rural_22 <dbl>, covid_vax100 <dbl>, who_region <fct>, iso_alpha3 <chr>miss_var_summary(ctry)# A tibble: 14 × 3
variable n_miss pct_miss
<chr> <int> <num>
1 births_attend 20 10.4
2 hale_all 9 4.69
3 hale_2000 9 4.69
4 covid_vax100 6 3.12
5 ihr_core_cap 2 1.04
6 wbank_income 1 0.521
7 c_num 0 0
8 country_name 0 0
9 univ_care 0 0
10 avg_tempc 0 0
11 female_22 0 0
12 rural_22 0 0
13 who_region 0 0
14 iso_alpha3 0 0 miss_case_table(ctry)# A tibble: 4 × 3
n_miss_in_case n_cases pct_cases
<int> <int> <dbl>
1 0 164 85.4
2 1 16 8.33
3 2 5 2.60
4 3 7 3.65- Outcome HALE_ALL
- 3 categorical variables (UNIV_CARE, WHO_REGION, WBANK_INCOME)
- 8 quantitative predictors (HALE_2000, BIRTHS_ATTEND, IHR_CORE_CAP, AVG_TEMPC, FEMALE_22, RURAL_22, COVID_VAX100)
and some missing values (at least in the predictors) to deal with…
22.3 Build single imputation
22.3.1 Filter out countries without information on our outcome, hale_all.
ctry_cc <- ctry |>
filter(complete.cases(hale_all))
pct_miss_case(ctry_cc)[1] 10.38251Later, when we do multiple imputation, we’ll run 15 imputations.
22.3.2 Single Imputation
Warning: Number of logged events: 322.3.3 Partition data after single imputation
We’ve seen other ways to partition the data. Here, let’s use data_partition() from the datawizard package, part of the easystats ecosystem.
22.4 Transforming the outcome?
fit0 <- lm(
hale_all ~ univ_care + wbank_income +
hale_2000 + births_attend + ihr_core_cap +
avg_tempc + female_22 + rural_22 + covid_vax100,
data = ctry_si_train
)boxCox(fit0)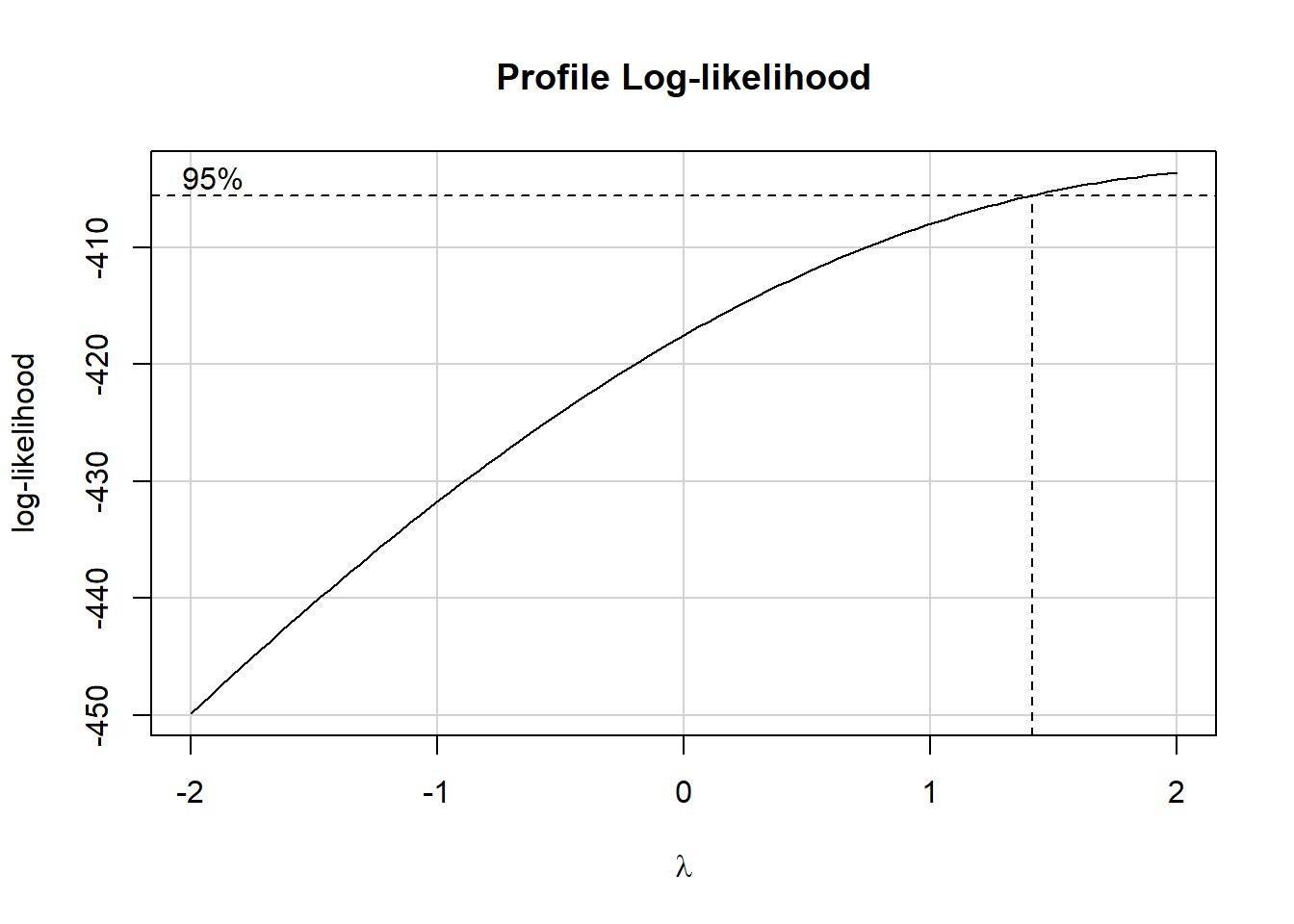
Let’s try the square of hale_all as our outcome.
p1 <- ggplot(ctry_si_train, aes(x = hale_all)) +
geom_histogram(bins = 20, fill = "coral", col = "blue")
p2 <- ggplot(ctry_si_train, aes(x = hale_all^2)) +
geom_histogram(bins = 20, fill = "aquamarine", col = "blue")
p1 + p2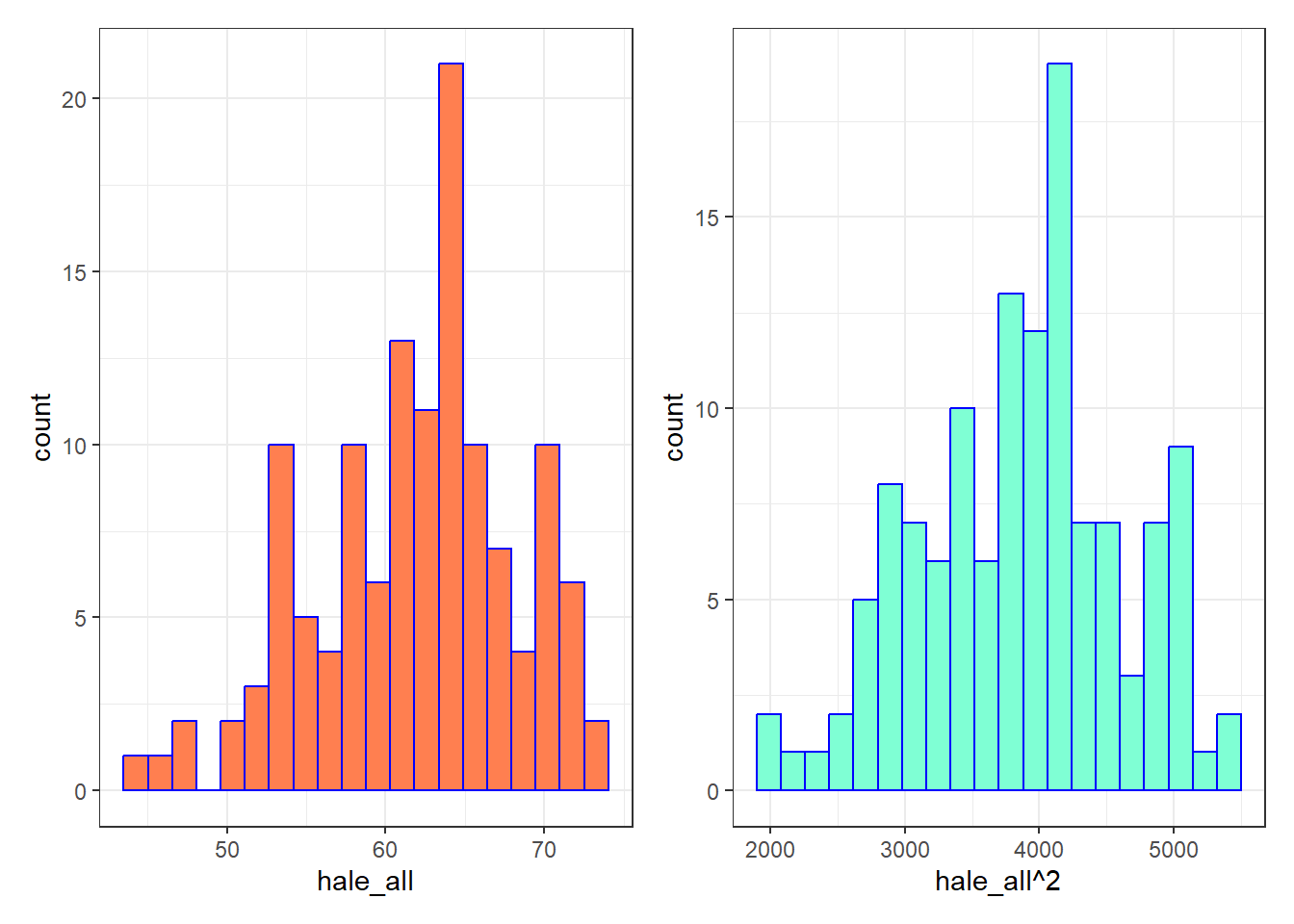
Not much of a difference, but perhaps we will see more meaningful improvements in our regression residual plots.
22.5 Fitting and Evaluating in Training Data
22.5.1 Kitchen Sink Model
fit1 <- lm( hale_sqr ~
univ_care + wbank_income + hale_2000 + births_attend +
ihr_core_cap + avg_tempc + female_22 + rural_22 +
covid_vax100,
data = ctry_si_train
)model_parameters(fit1)Parameter | Coefficient | SE | 95% CI | t(116) | p
------------------------------------------------------------------------------------------
(Intercept) | -291.03 | 598.75 | [-1476.93, 894.86] | -0.49 | 0.628
univ care [yes] | 44.22 | 61.01 | [ -76.62, 165.05] | 0.72 | 0.470
wbank income [Low] | -28.88 | 140.59 | [ -307.33, 249.58] | -0.21 | 0.838
wbank income [Lower-middle] | -186.50 | 98.98 | [ -382.54, 9.54] | -1.88 | 0.062
wbank income [Upper-middle] | -284.94 | 74.59 | [ -432.67, -137.21] | -3.82 | < .001
hale 2000 | 59.94 | 5.05 | [ 49.94, 69.94] | 11.87 | < .001
births attend | 4.81 | 2.11 | [ 0.64, 8.99] | 2.28 | 0.024
ihr core cap | 4.65 | 1.87 | [ 0.96, 8.35] | 2.49 | 0.014
avg tempc | -5.48 | 3.75 | [ -12.91, 1.95] | -1.46 | 0.147
female 22 | -2.05 | 9.07 | [ -20.01, 15.91] | -0.23 | 0.822
rural 22 | 1.67 | 1.62 | [ -1.54, 4.89] | 1.03 | 0.305
covid vax100 | 3.02 | 1.28 | [ 0.49, 5.56] | 2.36 | 0.020
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a Wald t-distribution approximation.model_performance(fit1)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
----------------------------------------------------------------------
1807.365 | 1810.558 | 1844.441 | 0.888 | 0.878 | 254.555 | 267.39822.5.2 LASSO to identify a subset
pred_x <- model.matrix(fit1)
out_y <- ctry_si_train |> select(hale_sqr) |> as.matrix()
set.seed(431)
lasso <- cv.glmnet(x = pred_x, y = out_y,
type.measure = "mse",
alpha = 1, family = "gaussian",
nlambda = 200, nfolds = 10)plot(lasso)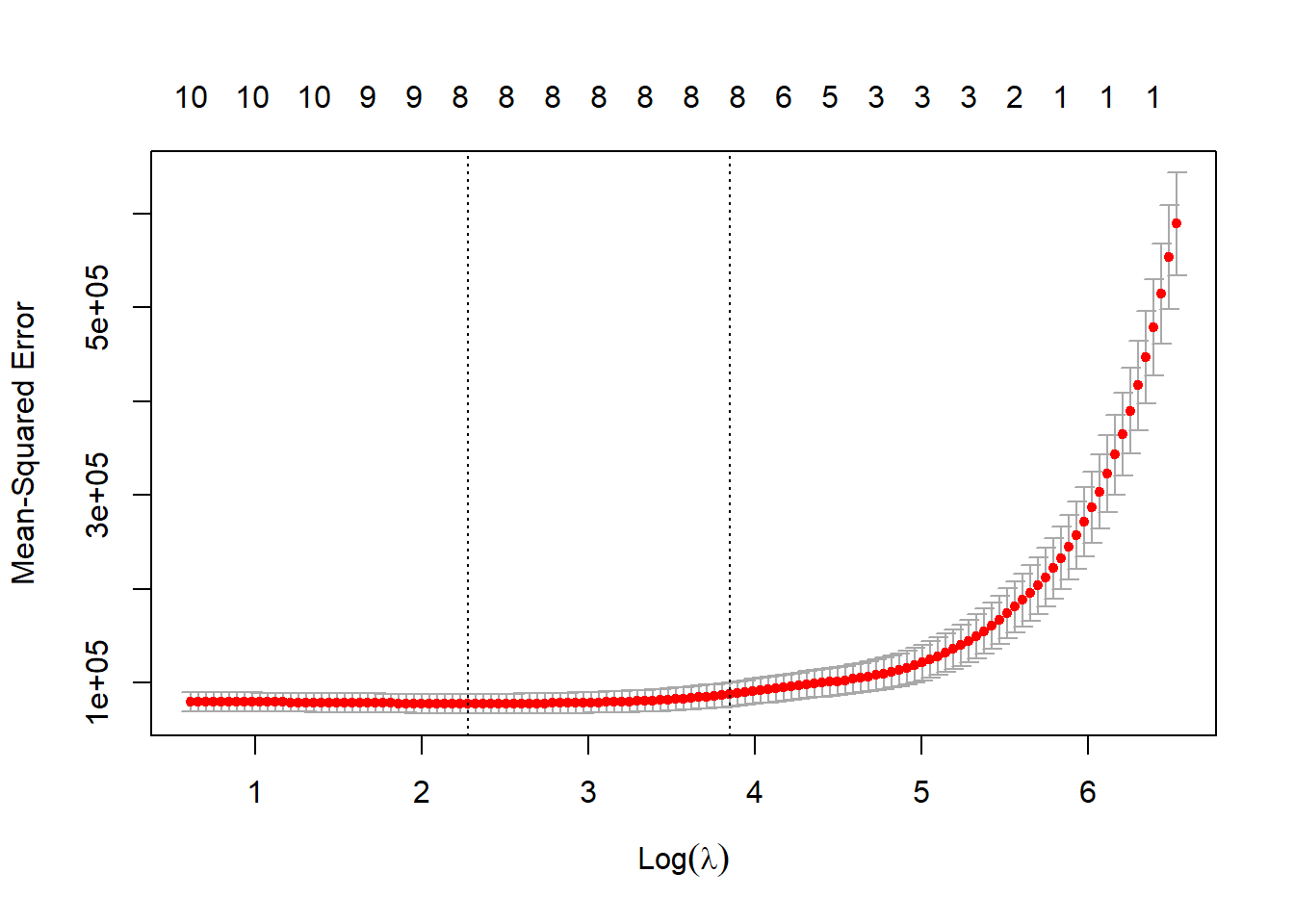
log(lasso$lambda.min)[1] 2.275831predict(lasso, type = "coef", s = "lambda.min")13 x 1 sparse Matrix of class "dgCMatrix"
lambda.min
(Intercept) -222.093827
(Intercept) .
univ_careyes 29.607576
wbank_incomeLow .
wbank_incomeLower-middle -129.930275
wbank_incomeUpper-middle -241.669217
hale_2000 58.555358
births_attend 4.299195
ihr_core_cap 4.520508
avg_tempc -4.936356
female_22 .
rural_22 .
covid_vax100 2.959860The LASSO suggestion is to drop two of the 9 variables in our original fit1 model, specifically female_22 and rural_22, since wbank_income is multi-categorical.
So that model would be:
fit7 <- lm( hale_sqr ~
univ_care + wbank_income + hale_2000 + births_attend +
ihr_core_cap + avg_tempc + covid_vax100,
data = ctry_si_train
)22.5.3 Stepwise Approach
fit6 <- select_parameters(fit1)
compare_models(fit1, fit6)Parameter | fit1 | fit6
--------------------------------------------------------------------------------------
(Intercept) | -291.03 (-1476.93, 894.86) | -288.36 (-992.56, 415.84)
wbank income [Low] | -28.88 ( -307.33, 249.58) | 1.23 (-263.48, 265.94)
wbank income [Lower-middle] | -186.50 ( -382.54, 9.54) | -165.91 (-343.22, 11.40)
wbank income [Upper-middle] | -284.94 ( -432.67, -137.21) | -280.72 (-421.53, -139.91)
hale 2000 | 59.94 ( 49.94, 69.94) | 59.16 ( 49.52, 68.80)
births attend | 4.81 ( 0.64, 8.99) | 4.96 ( 0.83, 9.10)
ihr core cap | 4.65 ( 0.96, 8.35) | 4.58 ( 1.08, 8.09)
avg tempc | -5.48 ( -12.91, 1.95) | -5.42 ( -11.86, 1.01)
covid vax100 | 3.02 ( 0.49, 5.56) | 3.08 ( 0.61, 5.55)
univ care [yes] | 44.22 ( -76.62, 165.05) |
rural 22 | 1.67 ( -1.54, 4.89) |
female 22 | -2.05 ( -20.01, 15.91) |
--------------------------------------------------------------------------------------
Observations | 128 | 128The stepwise approach drops three predictors, univ_care, rural_22 and female_22.
22.5.4 Best Subsets
How about using the best subsets approach?
k <- ols_step_best_subset(fit1,
max_order = 7,
metric = "cp")k Best Subsets Regression
------------------------------------------------------------------------------------------------
Model Index Predictors
------------------------------------------------------------------------------------------------
1 hale_2000
2 wbank_income hale_2000
3 wbank_income hale_2000 ihr_core_cap
4 wbank_income hale_2000 births_attend ihr_core_cap
5 wbank_income hale_2000 births_attend ihr_core_cap covid_vax100
6 wbank_income hale_2000 births_attend ihr_core_cap avg_tempc covid_vax100
7 wbank_income hale_2000 births_attend ihr_core_cap avg_tempc rural_22 covid_vax100
------------------------------------------------------------------------------------------------
Subsets Regression Summary
-------------------------------------------------------------------------------------------------------------------------------------------------
Adj. Pred
Model R-Square R-Square R-Square C(p) AIC SBIC SBC MSEP FPE HSP APC
-------------------------------------------------------------------------------------------------------------------------------------------------
1 0.8151 0.8136 0.8073 68.3017 1852.0658 1487.2564 1860.6218 13968177.2014 110831.2677 873.0094 0.1908
2 0.8556 0.8509 0.842 32.1908 1826.4294 1458.2510 1843.5416 10997356.3890 89354.3939 704.0977 0.1514
3 0.8724 0.8672 0.8573 16.6786 1812.5604 1445.0276 1832.5246 9794054.7936 80190.2350 632.1970 0.1358
4 0.8794 0.8734 0.8632 11.4137 1807.3525 1440.3096 1830.1687 9333656.2974 77004.7739 607.4578 0.1304
5 0.8844 0.8777 0.8668 8.1680 1803.8834 1437.4087 1829.5516 9017164.5402 74958.1159 591.7503 0.1269
6 0.8871 0.8795 0.8675 7.4172 1802.9193 1436.9166 1831.4396 8884176.1798 74408.9130 587.9223 0.1260
7 0.8879 0.8794 0.8666 8.5288 1803.9471 1438.2545 1835.3194 8891046.5054 75023.3563 593.3632 0.1270
-------------------------------------------------------------------------------------------------------------------------------------------------
AIC: Akaike Information Criteria
SBIC: Sawa's Bayesian Information Criteria
SBC: Schwarz Bayesian Criteria
MSEP: Estimated error of prediction, assuming multivariate normality
FPE: Final Prediction Error
HSP: Hocking's Sp
APC: Amemiya Prediction Criteria plot(k)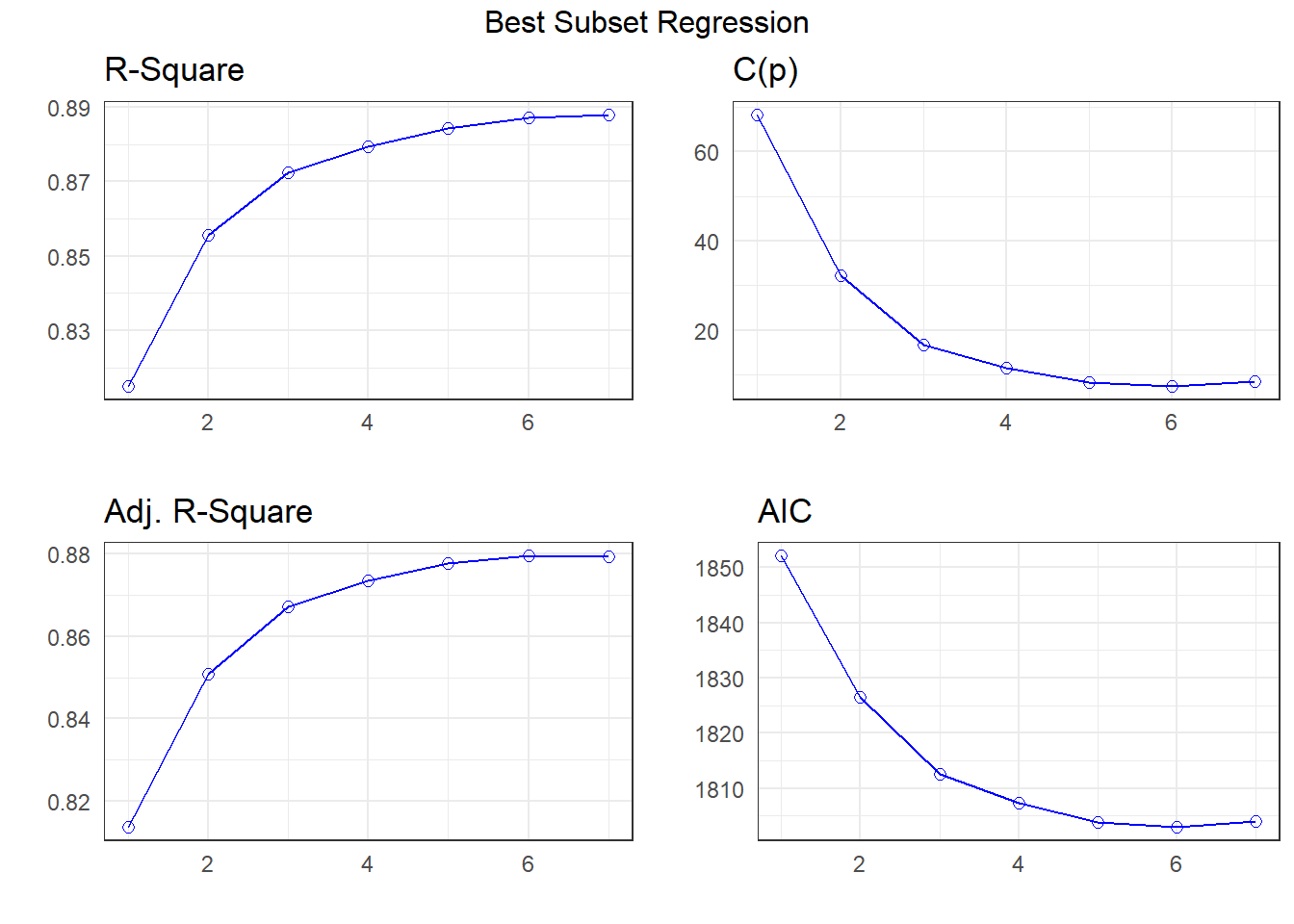
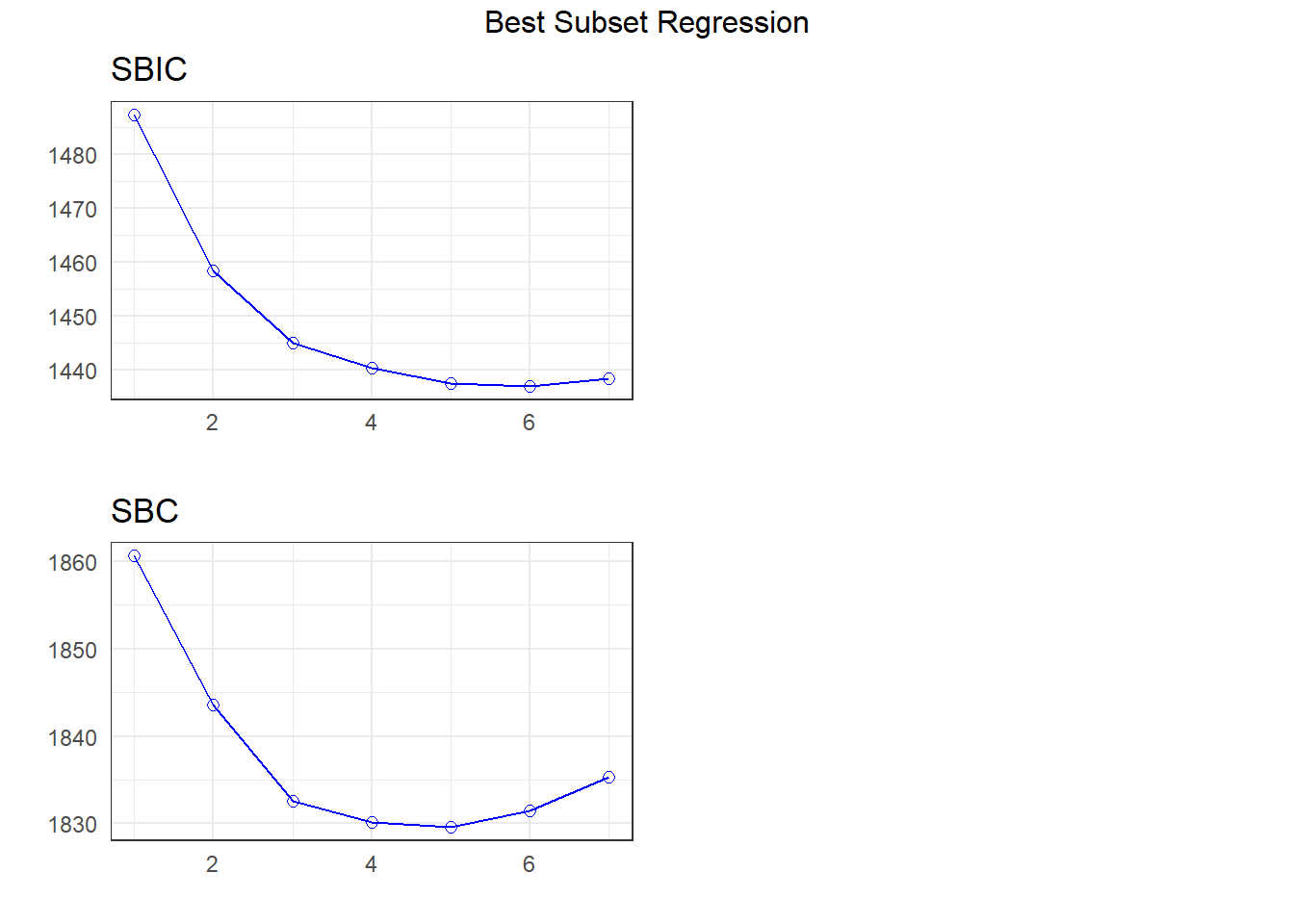
The best subsets suggestions include
- the same six-predictor model we got with stepwise
- a five-predictor model with
avg_tempcdropped from our 6-predictor model, and maybe - a different seven-parameter model than we obtained from the LASSO (keeping
rural_22but droppinguniv_care)
Let’s store the five-predictor model:
fit5 <- lm( hale_sqr ~
wbank_income + hale_2000 + births_attend +
ihr_core_cap + covid_vax100,
data = ctry_si_train
)22.5.5 Compare Candidate Models in Training Sample
So we have at least four candidate models: fit1, fit5, fit6 and fit7.
compare_models(fit1, fit7, fit6, fit5)Parameter | fit1 | fit7 | fit6 | fit5
-------------------------------------------------------------------------------------------------------------------------------------------------
(Intercept) | -291.03 (-1476.93, 894.86) | -263.05 (-974.18, 448.09) | -288.36 (-992.56, 415.84) | -439.16 (-1125.27, 246.94)
wbank income [Low] | -28.88 ( -307.33, 249.58) | 1.60 (-263.85, 267.06) | 1.23 (-263.48, 265.94) | -33.50 ( -296.90, 229.91)
wbank income [Lower-middle] | -186.50 ( -382.54, 9.54) | -157.94 (-337.70, 21.81) | -165.91 (-343.22, 11.40) | -194.85 ( -370.09, -19.62)
wbank income [Upper-middle] | -284.94 ( -432.67, -137.21) | -279.75 (-420.99, -138.52) | -280.72 (-421.53, -139.91) | -300.61 ( -440.45, -160.78)
hale 2000 | 59.94 ( 49.94, 69.94) | 58.86 ( 49.13, 68.58) | 59.16 ( 49.52, 68.80) | 59.49 ( 49.78, 69.19)
births attend | 4.81 ( 0.64, 8.99) | 4.87 ( 0.71, 9.02) | 4.96 ( 0.83, 9.10) | 5.29 ( 1.15, 9.44)
ihr core cap | 4.65 ( 0.96, 8.35) | 4.42 ( 0.87, 7.98) | 4.58 ( 1.08, 8.09) | 5.04 ( 1.55, 8.53)
covid vax100 | 3.02 ( 0.49, 5.56) | 3.00 ( 0.50, 5.49) | 3.08 ( 0.61, 5.55) | 2.86 ( 0.39, 5.34)
univ care [yes] | 44.22 ( -76.62, 165.05) | 35.15 ( -81.32, 151.62) | |
female 22 | -2.05 ( -20.01, 15.91) | | |
avg tempc | -5.48 ( -12.91, 1.95) | -5.35 ( -11.81, 1.10) | -5.42 ( -11.86, 1.01) |
rural 22 | 1.67 ( -1.54, 4.89) | | |
-------------------------------------------------------------------------------------------------------------------------------------------------
Observations | 128 | 128 | 128 | 128compare_performance(fit1, fit7, fit6, fit5)# Comparison of Model Performance Indices
Name | Model | AIC (weights) | AICc (weights) | BIC (weights) | R2 | R2 (adj.) | RMSE | Sigma
-------------------------------------------------------------------------------------------------------
fit1 | lm | 1807.4 (0.050) | 1810.6 (0.026) | 1844.4 (<.001) | 0.888 | 0.878 | 254.555 | 267.398
fit7 | lm | 1804.5 (0.206) | 1806.8 (0.170) | 1835.9 (0.029) | 0.887 | 0.879 | 255.719 | 266.334
fit6 | lm | 1802.9 (0.460) | 1804.8 (0.463) | 1831.4 (0.272) | 0.887 | 0.879 | 256.106 | 265.614
fit5 | lm | 1803.9 (0.284) | 1805.4 (0.341) | 1829.6 (0.699) | 0.884 | 0.878 | 259.088 | 267.585plot(compare_performance(fit1, fit7, fit6, fit5))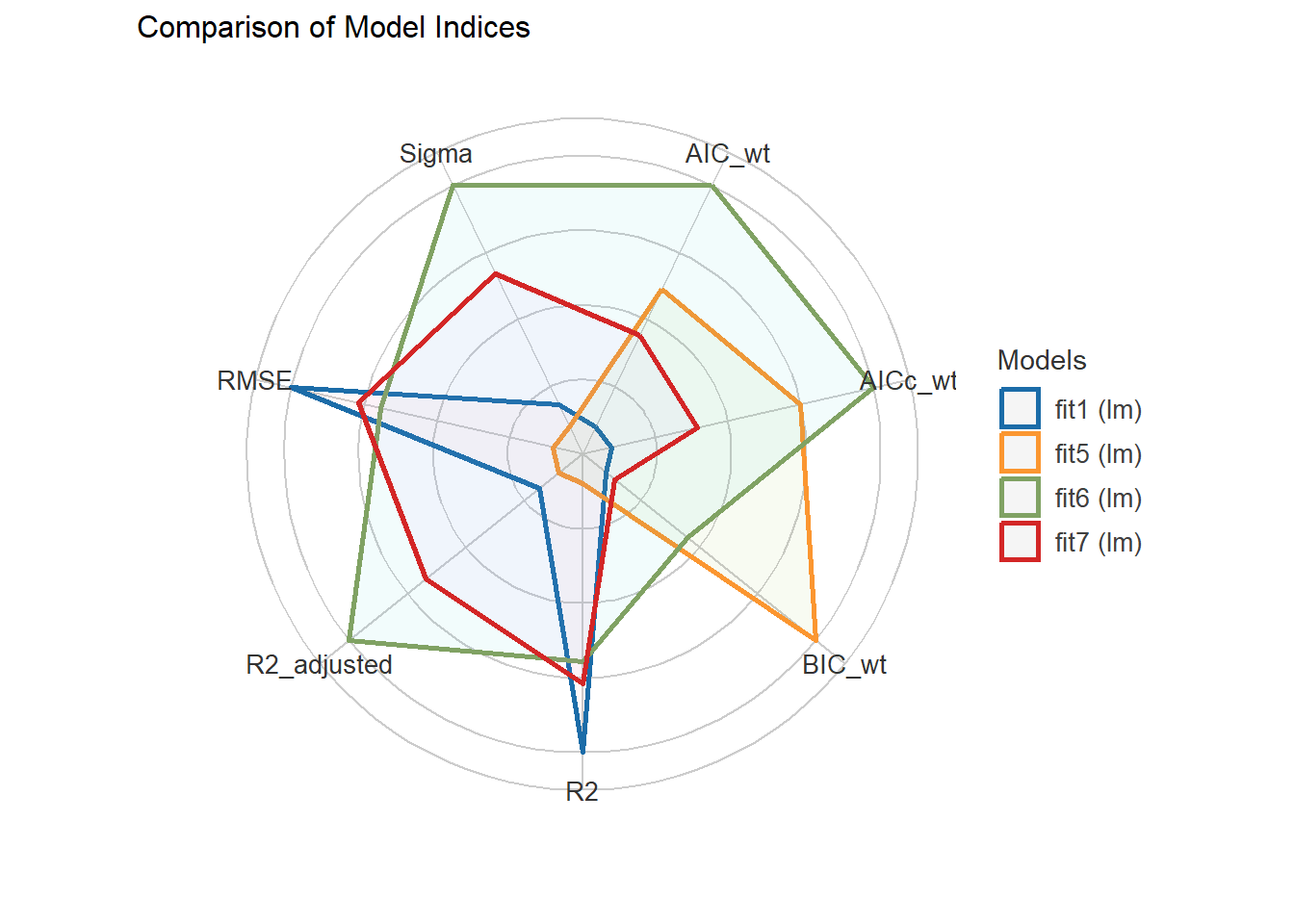
It looks like the six-predictor model is the best choice (but just barely) using adjusted \(R^2\), \(\sigma\) and AIC. Let’s look at the test sample.
22.5.6 Compare Candidate Models in Test Sample
test1 <- augment(fit1, newdata = ctry_si_test) |>
mutate(mod_n = "Model 1 (KS)")
test7 <- augment(fit7, newdata = ctry_si_test) |>
mutate(mod_n = "Model 7")
test6 <- augment(fit6, newdata = ctry_si_test) |>
mutate(mod_n = "Model 6")
test5 <- augment(fit5, newdata = ctry_si_test) |>
mutate(mod_n = "Model 5")
test_res <- bind_rows(test1, test7, test6, test5) |>
mutate(predicted_haleall = sqrt(.fitted),
resid_haleall = hale_all - predicted_haleall) |>
select(mod_n, country_name, hale_all,
predicted_haleall, resid_haleall, everything()) |>
arrange(c_num, mod_n)head(test_res, 8)# A tibble: 8 × 21
mod_n country_name hale_all predicted_haleall resid_haleall c_num univ_care
<chr> <chr> <dbl> <dbl> <dbl> <chr> <fct>
1 Model 1… Luxembourg 71.2 68.6 2.55 100 yes
2 Model 5 Luxembourg 71.2 68.5 2.68 100 yes
3 Model 6 Luxembourg 71.2 68.7 2.49 100 yes
4 Model 7 Luxembourg 71.2 68.8 2.39 100 yes
5 Model 1… Maldives 66.7 63.8 2.90 104 yes
6 Model 5 Maldives 66.7 63.6 3.15 104 yes
7 Model 6 Maldives 66.7 63.2 3.53 104 yes
8 Model 7 Maldives 66.7 63.3 3.38 104 yes
# ℹ 14 more variables: wbank_income <fct>, hale_2000 <dbl>,
# births_attend <dbl>, ihr_core_cap <dbl>, avg_tempc <dbl>, female_22 <dbl>,
# rural_22 <dbl>, covid_vax100 <dbl>, who_region <fct>, iso_alpha3 <chr>,
# .row_id <int>, hale_sqr <dbl>, .fitted <dbl>, .resid <dbl>test_res |>
group_by(mod_n) |>
summarise(MAPE = mean(abs(resid_haleall)),
medAPE = median(abs(resid_haleall)),
maxAPE = max(abs(resid_haleall)),
RMSPE = sqrt(mean(resid_haleall^2)),
rsqr = cor(hale_all, predicted_haleall)^2) |>
kable()| mod_n | MAPE | medAPE | maxAPE | RMSPE | rsqr |
|---|---|---|---|---|---|
| Model 1 (KS) | 1.399561 | 1.172160 | 4.242786 | 1.682548 | 0.9063445 |
| Model 5 | 1.387650 | 1.091875 | 3.818286 | 1.700453 | 0.9009602 |
| Model 6 | 1.357642 | 1.198238 | 4.019098 | 1.660509 | 0.9072666 |
| Model 7 | 1.347411 | 1.168969 | 3.973297 | 1.637438 | 0.9092709 |
We see that:
- Model 6 has the smallest mean, median and maximum average prediction error, but
- Model 7 has the smallest RMSPE and largest validated \(R^2\).
I’ll select model 6, assuming that model assumptions aren’t too badly violated in our training sample. Let’s check that.
22.6 Check Assumptions in Winning Model
check_model(fit6)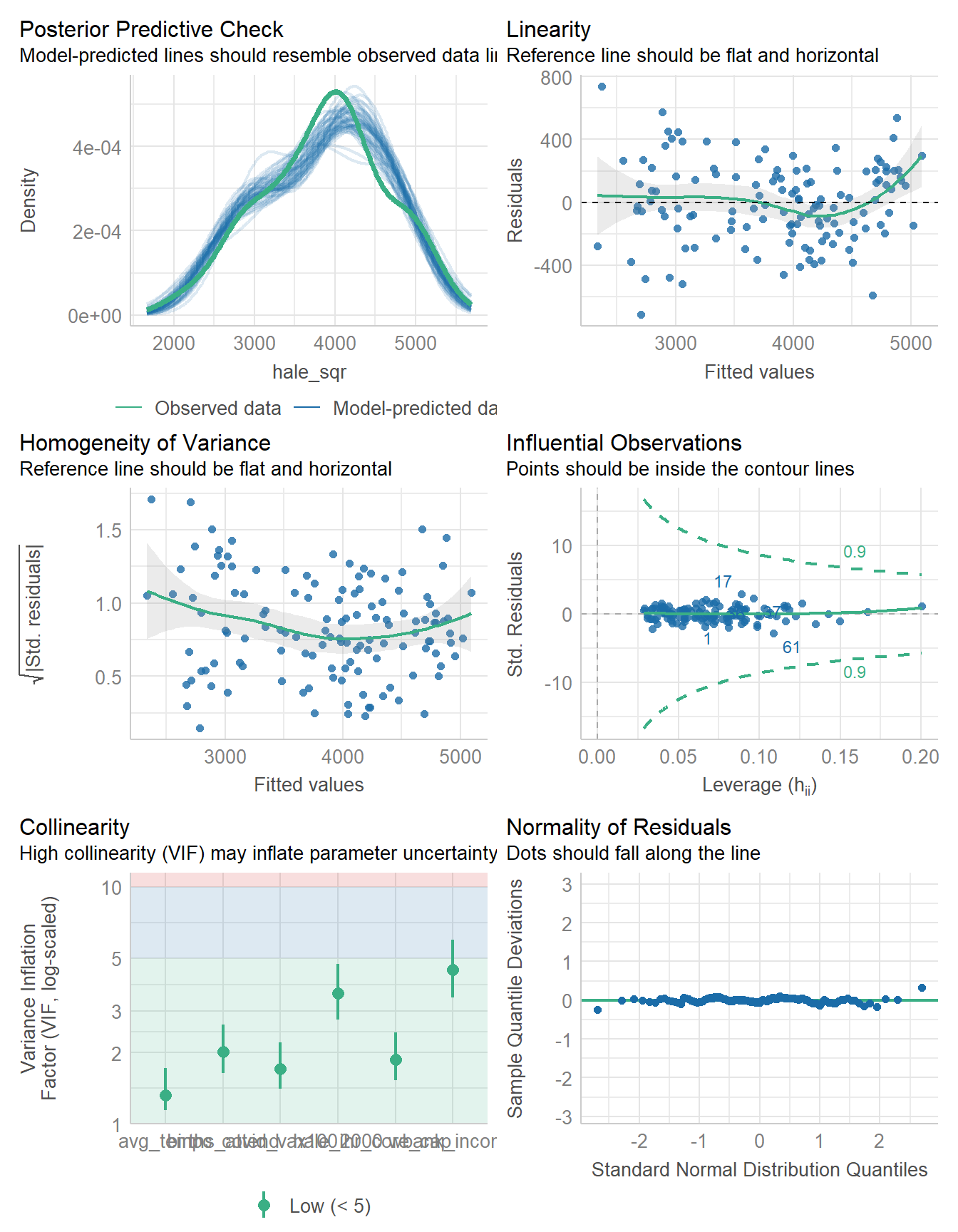
The collinearity is OK now, and outside of a couple of observations, I think Normality isn’t a serious problem.
22.7 Estimates for the Winning Model
So now, I’ll refit the six-predictor model, which includes:
wbank_incomehale_2000births_attendihr_core_cap-
avg_tempc, and covid_vax100
to the entire set of data, with various assumptions about the missing data.
22.7.1 Complete Case Analysis
Here, we’ll fit the model to all of the data except (as we’ll do in all three cases) the countries without outcome (hale_all) information.
fit_cc <- lm((hale_all^2) ~ wbank_income + hale_2000 +
births_attend + ihr_core_cap +
avg_tempc + covid_vax100,
data = ctry_cc)
model_parameters(fit_cc, ci = 0.95)Parameter | Coefficient | SE | 95% CI | t(155) | p
-----------------------------------------------------------------------------------------
(Intercept) | -93.51 | 302.42 | [-690.90, 503.89] | -0.31 | 0.758
wbank income [Low] | -68.39 | 112.63 | [-290.88, 154.11] | -0.61 | 0.545
wbank income [Lower-middle] | -148.19 | 73.52 | [-293.42, -2.97] | -2.02 | 0.046
wbank income [Upper-middle] | -267.91 | 60.30 | [-387.02, -148.81] | -4.44 | < .001
hale 2000 | 57.44 | 4.20 | [ 49.14, 65.74] | 13.67 | < .001
births attend | 4.84 | 1.79 | [ 1.31, 8.37] | 2.71 | 0.008
ihr core cap | 3.80 | 1.51 | [ 0.81, 6.79] | 2.51 | 0.013
avg tempc | -5.51 | 2.80 | [ -11.05, 0.03] | -1.97 | 0.051
covid vax100 | 2.65 | 1.09 | [ 0.49, 4.81] | 2.42 | 0.017
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a Wald t-distribution approximation.model_performance(fit_cc)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
-------------------------------------------------------------------------
44110.965 | 44112.403 | 44141.963 | 0.881 | 0.874 | 247.017 | 254.08822.7.2 After Single Imputation
fit_si <- lm((hale_all^2) ~ wbank_income + hale_2000 +
births_attend + ihr_core_cap +
avg_tempc + covid_vax100,
data = ctry_si)
model_parameters(fit_si, ci = 0.95)Parameter | Coefficient | SE | 95% CI | t(174) | p
-----------------------------------------------------------------------------------------
(Intercept) | -49.82 | 270.59 | [-583.88, 484.25] | -0.18 | 0.854
wbank income [Low] | -79.91 | 97.08 | [-271.51, 111.69] | -0.82 | 0.412
wbank income [Lower-middle] | -178.92 | 65.89 | [-308.97, -48.86] | -2.72 | 0.007
wbank income [Upper-middle] | -293.54 | 56.08 | [-404.22, -182.87] | -5.23 | < .001
hale 2000 | 57.96 | 3.67 | [ 50.72, 65.20] | 15.80 | < .001
births attend | 4.50 | 1.63 | [ 1.29, 7.72] | 2.76 | 0.006
ihr core cap | 3.75 | 1.41 | [ 0.97, 6.53] | 2.66 | 0.008
avg tempc | -6.47 | 2.60 | [ -11.60, -1.35] | -2.49 | 0.014
covid vax100 | 2.64 | 0.98 | [ 0.70, 4.57] | 2.69 | 0.008
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a Wald t-distribution approximation.model_performance(fit_si)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
-------------------------------------------------------------------------
52919.799 | 52921.078 | 52951.894 | 0.892 | 0.887 | 239.530 | 245.64722.7.3 Multiple Imputation
As mentioned earlier, I am dropping the observations with no information on my outcome (hale_all) which is the ctry_cc data. Then I am building 15 imputations, because I am missing data on just over 10% of cases within ctry_cc.
pct_miss_case(ctry_cc) # as a reminder[1] 10.38251Now, we fit our six-predictor model in each of these 15 imputed data sets, like this:
imp_ests <- ctry_cc |>
mice(m = 15, seed = 431, print = FALSE) |>
with(lm((hale_all)^2 ~ wbank_income + hale_2000 +
births_attend + ihr_core_cap +
avg_tempc + covid_vax100)) |>
pool()Warning: Number of logged events: 3model_parameters(imp_ests, ci = 0.95)Warning: Number of logged events: 3
Warning: Number of logged events: 3
Warning: Number of logged events: 3
Warning: Number of logged events: 3# Fixed Effects
Parameter | Coefficient | SE | 95% CI | t | df | p
-------------------------------------------------------------------------------------------------
(Intercept) | -66.57 | 274.03 | [-607.55, 474.42] | -0.24 | 168.60 | 0.808
wbank income [Low] | -74.63 | 97.66 | [-267.42, 118.15] | -0.76 | 169.38 | 0.446
wbank income [Lower-middle] | -172.29 | 66.57 | [-303.70, -40.88] | -2.59 | 170.49 | 0.010
wbank income [Upper-middle] | -294.49 | 56.44 | [-405.89, -183.09] | -5.22 | 171.96 | < .001
hale 2000 | 58.20 | 3.69 | [ 50.91, 65.49] | 15.76 | 169.20 | < .001
births attend | 4.50 | 1.66 | [ 1.23, 7.77] | 2.72 | 166.98 | 0.007
ihr core cap | 3.70 | 1.42 | [ 0.91, 6.50] | 2.62 | 171.40 | 0.010
avg tempc | -6.47 | 2.61 | [ -11.62, -1.32] | -2.48 | 171.81 | 0.014
covid vax100 | 2.70 | 1.01 | [ 0.72, 4.68] | 2.69 | 171.28 | 0.008
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a Wald t-distribution approximation.glance(imp_ests) nimp nobs r.squared adj.r.squared
1 15 183 0.8915267 0.8865394and hopefully show them all to be fairly similar. Then describe that winning fit in some detail, probably in the multiple imputation case.
22.8 Bayesian Linear Fit
Show Bayesian fit for the winning model across the entire sample, and draw conclusions about its fit and form.
ctry_si <- ctry_si |>
mutate(hale_sqr = hale_all^2)
fit_bay <- stan_glm(hale_sqr ~
wbank_income + hale_2000 +
births_attend + ihr_core_cap +
avg_tempc + covid_vax100,
data = ctry_si, refresh = 0)
model_parameters(fit_bay, ci = 0.95)Parameter | Median | 95% CI | pd | Rhat | ESS | Prior
----------------------------------------------------------------------------------------------------------------
(Intercept) | -58.22 | [-593.32, 491.26] | 58.45% | 1.000 | 2586.00 | Normal (3859.71 +- 1825.71)
wbank_incomeLow | -79.75 | [-272.32, 116.58] | 78.38% | 1.001 | 2311.00 | Normal (0.00 +- 5057.64)
wbank_incomeLower-middle | -177.89 | [-309.74, -47.08] | 99.50% | 1.000 | 2335.00 | Normal (0.00 +- 4014.05)
wbank_incomeUpper-middle | -292.57 | [-409.28, -179.50] | 100% | 1.001 | 3126.00 | Normal (0.00 +- 4111.90)
hale_2000 | 58.05 | [ 50.54, 64.89] | 100% | 1.001 | 3029.00 | Normal (0.00 +- 209.73)
births_attend | 4.54 | [ 1.29, 7.67] | 99.72% | 1.000 | 4459.00 | Normal (0.00 +- 113.08)
ihr_core_cap | 3.74 | [ 0.99, 6.54] | 99.55% | 1.000 | 4377.00 | Normal (0.00 +- 101.06)
avg_tempc | -6.43 | [ -11.52, -1.41] | 99.33% | 1.000 | 4533.00 | Normal (0.00 +- 225.51)
covid_vax100 | 2.60 | [ 0.68, 4.50] | 99.62% | 1.000 | 4575.00 | Normal (0.00 +- 74.26)
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a MCMC distribution approximation.model_performance(fit_bay)# Indices of model performance
ELPD | ELPD_SE | LOOIC | LOOIC_SE | WAIC | R2 | R2 (adj.) | RMSE | Sigma
--------------------------------------------------------------------------------------------
-1272.992 | 10.426 | 2545.984 | 20.853 | 2545.895 | 0.887 | 0.880 | 239.536 | 246.89722.9 For More Information
point to https://r4ds.hadley.nz/missing-values