11 Association and Regression
11.1 R setup for this chapter
Appendix A lists all R packages used in this book, and also provides R session information. Appendix B describes the 431-Love.R script, and demonstrates its use.
11.2 Data: US Wooden Roller Coasters
Appendix C provides further guidance on pulling data from other systems into R, while Appendix D gives more information (including download links) for all data sets used in this book.
I was motivated to look at roller coaster1 data by an older example from the Data and Story Library which also uses the Roller Coaster Database. Specifically, I used this URL to pull data on 110 currently operating (as of late June 2024) wooden roller coasters in the US meeting the following criteria:
- Classification = Roller Coaster
- Design = Sit Down
- Existing
- Location = United States
- Roller Coasters
- Current Status = Operating
- Type = Wood
coasters <- read_csv("data/coasters.csv", show_col_types = FALSE) |>
janitor::clean_names()
head(coasters)# A tibble: 6 × 10
c_id coaster_n speed height duration opened park town state region
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr>
1 C-001 American Eagle 66 127 143 1981 Six … Gurn… Illi… Midwe…
2 C-002 American Thunder 48 82 NA 2008 Six … Eure… Miss… Midwe…
3 C-003 Apocalypse the Ri… 50.1 95 180 2009 Six … Vale… Cali… West
4 C-004 Arkansas Twister NA 95 NA 1992 Magi… Hot … Arka… South
5 C-005 Beast 64.8 110 250 1979 King… Mason Ohio Midwe…
6 C-006 Big Dipper NA NA 100 1958 Camd… Hunt… West… South The 10 variables in the data are:
| Variable | Description |
|---|---|
c_ID |
Coaster Identification Code |
coaster_N |
Name of Coaster |
speed |
Maximum speed, in miles per hour |
height |
Maximum height, in feet |
duration |
Length of ride, in seconds |
opened |
Year of opening |
park |
Name of amusement park |
town |
Name of town where park is located |
state |
State where park is located |
region |
Region of the US (4 categories) |
11.2.1 Missing Data
The naniar package provides some excellent ways to count missing values in our data.
miss_var_summary(coasters)# A tibble: 10 × 3
variable n_miss pct_miss
<chr> <int> <num>
1 duration 35 31.8
2 speed 17 15.5
3 height 9 8.18
4 c_id 0 0
5 coaster_n 0 0
6 opened 0 0
7 park 0 0
8 town 0 0
9 state 0 0
10 region 0 0 miss_case_table(coasters)# A tibble: 4 × 3
n_miss_in_case n_cases pct_cases
<int> <int> <dbl>
1 0 64 58.2
2 1 32 29.1
3 2 13 11.8
4 3 1 0.909As we’ve done in the past, we’ll restrict ourselves to just the 64 coasters with complete data on all variables. We’ll also set up the ages of the coasters, where age is just 2024 minus the year the coaster opened.
# A tibble: 64 × 11
c_id coaster_n speed height duration opened park town state region age
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl>
1 C-001 American E… 66 127 143 1981 Six … Gurn… Illi… Midwe… 43
2 C-003 Apocalypse… 50.1 95 180 2009 Six … Vale… Cali… West 15
3 C-005 Beast 64.8 110 250 1979 King… Mason Ohio Midwe… 45
4 C-007 Blue Streak 40 78 105 1964 Ceda… Sand… Ohio Midwe… 60
5 C-013 Classic Co… 50 55 105 1935 Wash… Puya… Wash… West 89
6 C-014 Coastersau… 32 40 50 2004 Lego… Wint… Flor… South 20
7 C-015 Comet 50 84 105 1946 Hers… Hers… Penn… North… 78
8 C-016 Comet 55 95 120 1994 Six … Quee… New … North… 30
9 C-017 Comet 25 37 84 1951 Wald… Erie Penn… North… 73
10 C-019 Cyclone 60 75 110 1927 Luna… Broo… New … North… 97
# ℹ 54 more rows11.3 Exploratory Data Analysis
11.3.1 Visualizations
Although we’re primarily interested in associations between variables, let’s look briefly at the sample distributions for the four variables of interest.
p1 <- ggplot(coast_cc, aes(x = speed)) +
geom_histogram(bins = 8, fill = "red", col = "yellow") +
labs(title = "Speed in MPH")
p2 <- ggplot(coast_cc, aes(x = height)) +
geom_histogram(bins = 8, fill = "slateblue", col = "yellow") +
labs(title = "Height in feet")
p3 <- ggplot(coast_cc, aes(x = duration)) +
geom_histogram(bins = 8, fill = "magenta", col = "yellow") +
labs(title = "Duration in seconds")
p4 <- ggplot(coast_cc, aes(x = age)) +
geom_histogram(bins = 8, fill = "seagreen", col = "yellow") +
labs(title = "Age in years")
(p1 + p2) / (p3 + p4) +
plot_annotation(title = "Roller Coaster Data: Quantities",
subtitle = "Histograms, n = 64 coasters")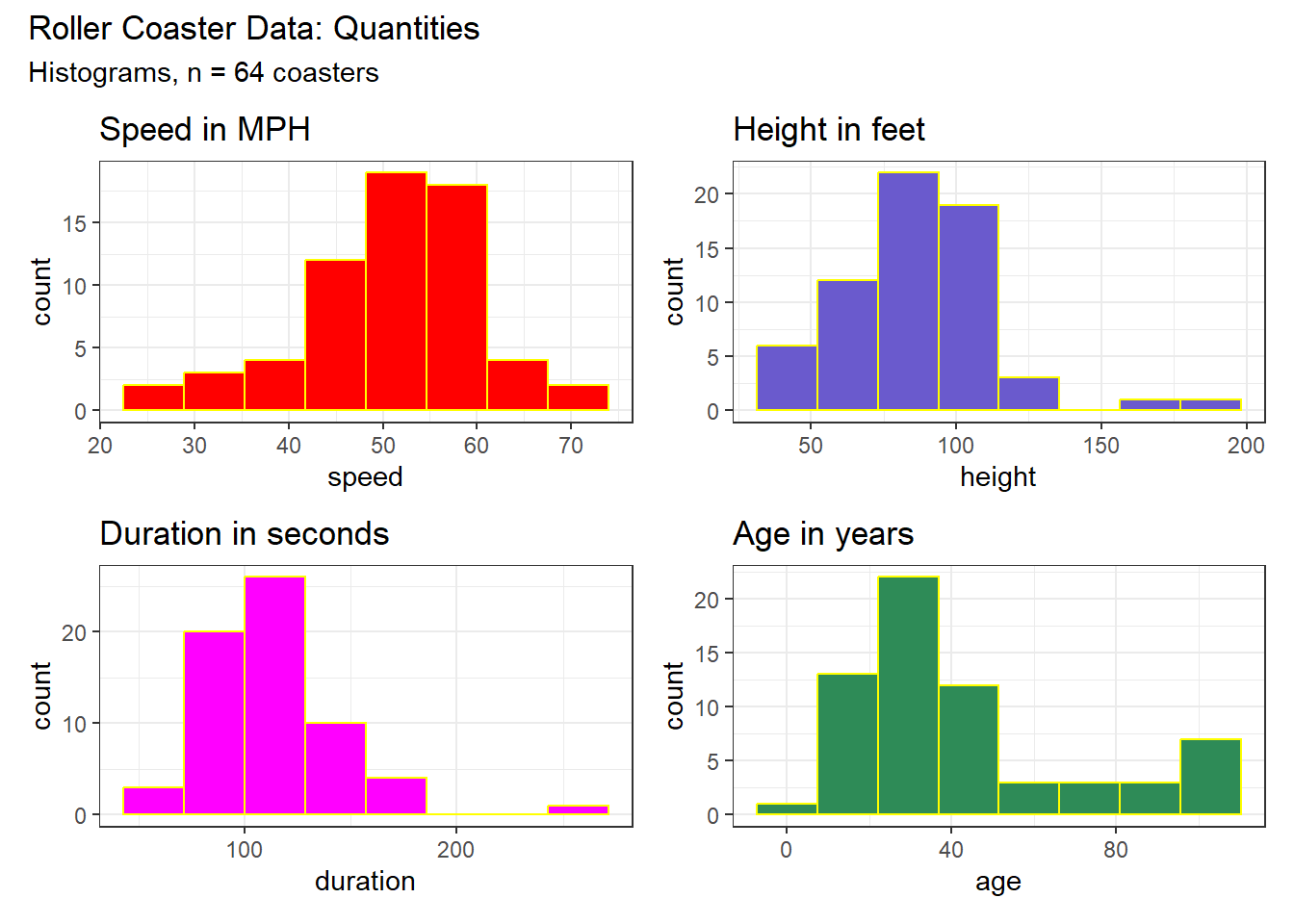
11.3.2 Numeric Summaries
Here are distributional descriptions of our four main variables of interest.
res1 <- coast_cc |>
reframe(lovedist(speed)) |>
mutate(variable = "speed")
res2 <- coast_cc |>
reframe(lovedist(height)) |>
mutate(variable = "height")
res3 <- coast_cc |>
reframe(lovedist(duration)) |>
mutate(variable = "duration")
res4 <- coast_cc |>
reframe(lovedist(age)) |>
mutate(variable = "age")
rbind(res1, res2, res3, res4) |>
relocate(variable) |>
kable(digits = 2)| variable | n | miss | mean | sd | med | mad | min | q25 | q75 | max |
|---|---|---|---|---|---|---|---|---|---|---|
| speed | 64 | 0 | 50.76 | 9.06 | 51.15 | 7.19 | 25 | 45.75 | 56.00 | 70 |
| height | 64 | 0 | 86.20 | 26.19 | 85.00 | 22.24 | 35 | 70.75 | 100.00 | 181 |
| duration | 64 | 0 | 112.67 | 31.71 | 111.00 | 28.17 | 50 | 90.00 | 120.00 | 250 |
| age | 64 | 0 | 42.72 | 27.57 | 33.50 | 20.76 | 1 | 25.00 | 51.25 | 104 |
11.4 height - speed association
First, we’ll consider the association of each coaster’s speed with its height.
11.4.1 Scatterplot with Pearson correlation
Let’s create a plot including both the linear fit (from lm()) and a loess smooth, as well as the Pearson correlation coefficient.
ggplot(coast_cc, aes(x = height, y = speed)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x, col = "red") +
geom_smooth(method = "loess", formula = y ~ x, se = FALSE, col = "blue") +
labs(title = "Association of Speed with Height") +
annotate("text", x = 150, y = 40,
label = glue("Pearson r = ",
round(cor(coast_cc$height,
coast_cc$speed),3)))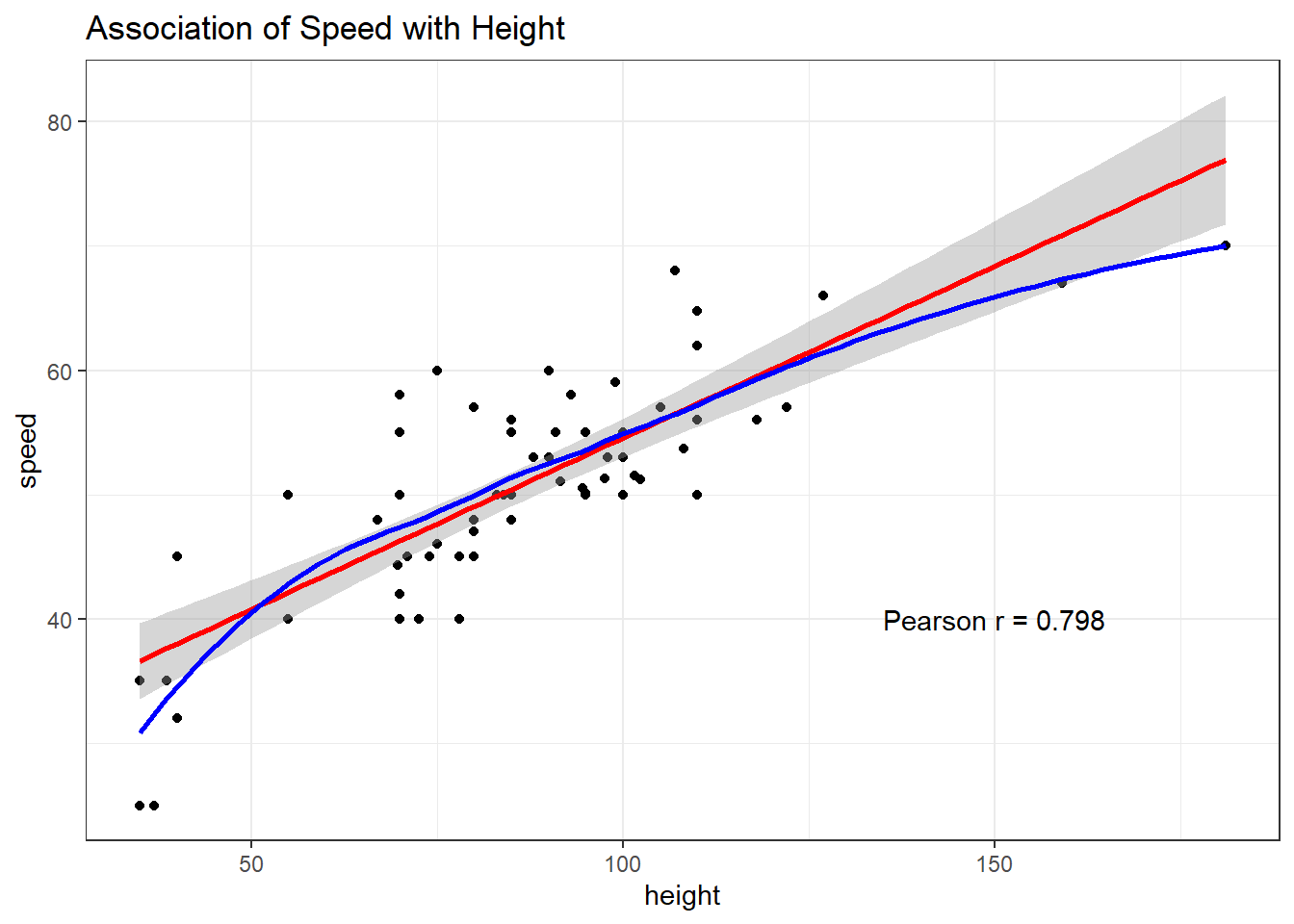
Another way to augment the scatterplot with similar information (other than the loess smooth) follows:
res6 <- cor_test(coast_cc, "height", "speed")
res6Parameter1 | Parameter2 | r | 95% CI | t(62) | p
-----------------------------------------------------------------
height | speed | 0.80 | [0.69, 0.87] | 10.43 | < .001***
Observations: 64plot(res6)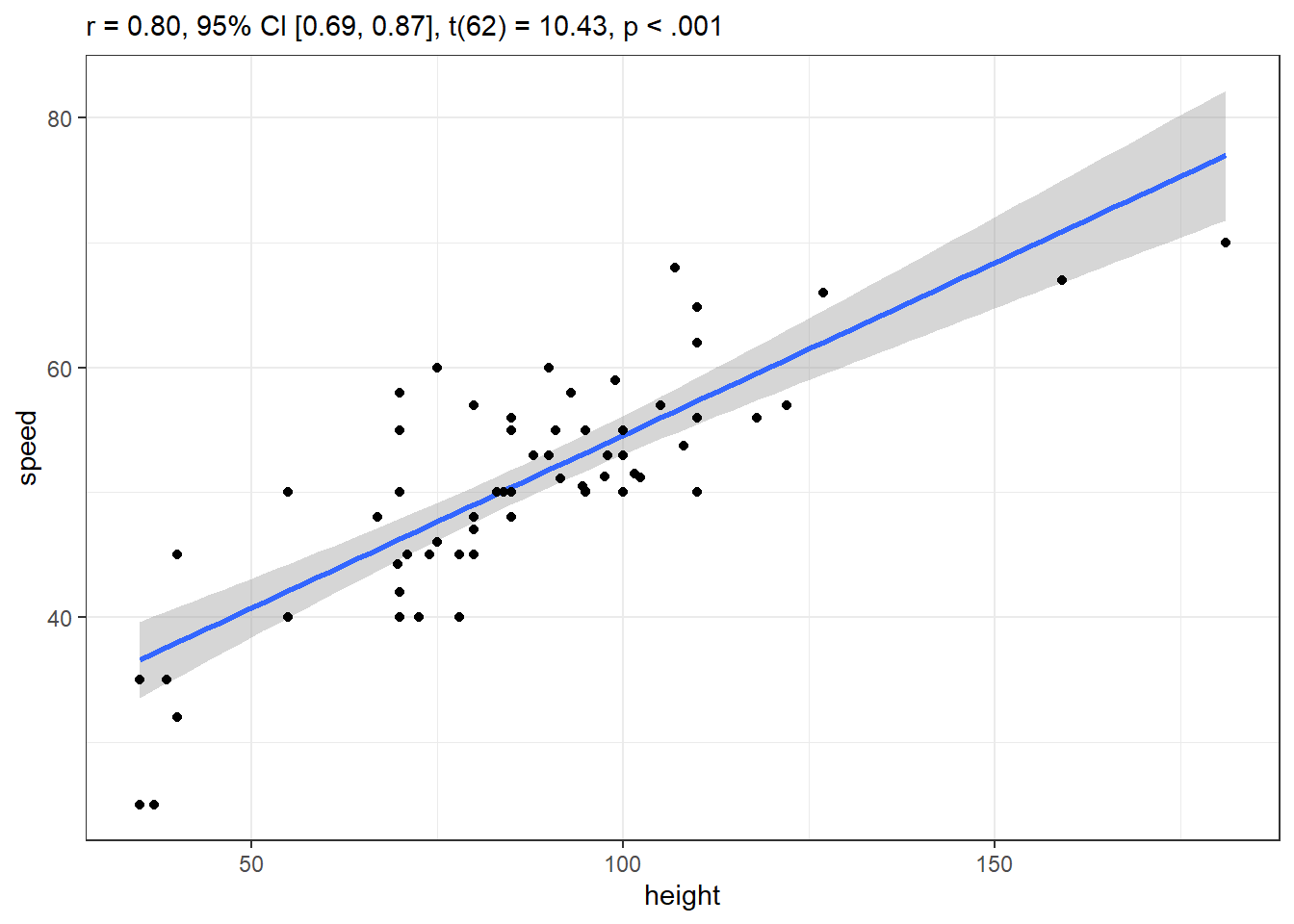
11.4.2 Spearman’s rank correlation
This measure is the Pearson correlation of the rank scores of the two variables. It’s mostly used for assessing whether or not an association is monotone.
cor_test(coast_cc, "height", "speed", method = "spearman")Parameter1 | Parameter2 | rho | 95% CI | S | p
--------------------------------------------------------------------
height | speed | 0.72 | [0.57, 0.82] | 12371.84 | < .001***
Observations: 64Numerous other measures of association are available. See this description, which is part of easystats’ correlation package.
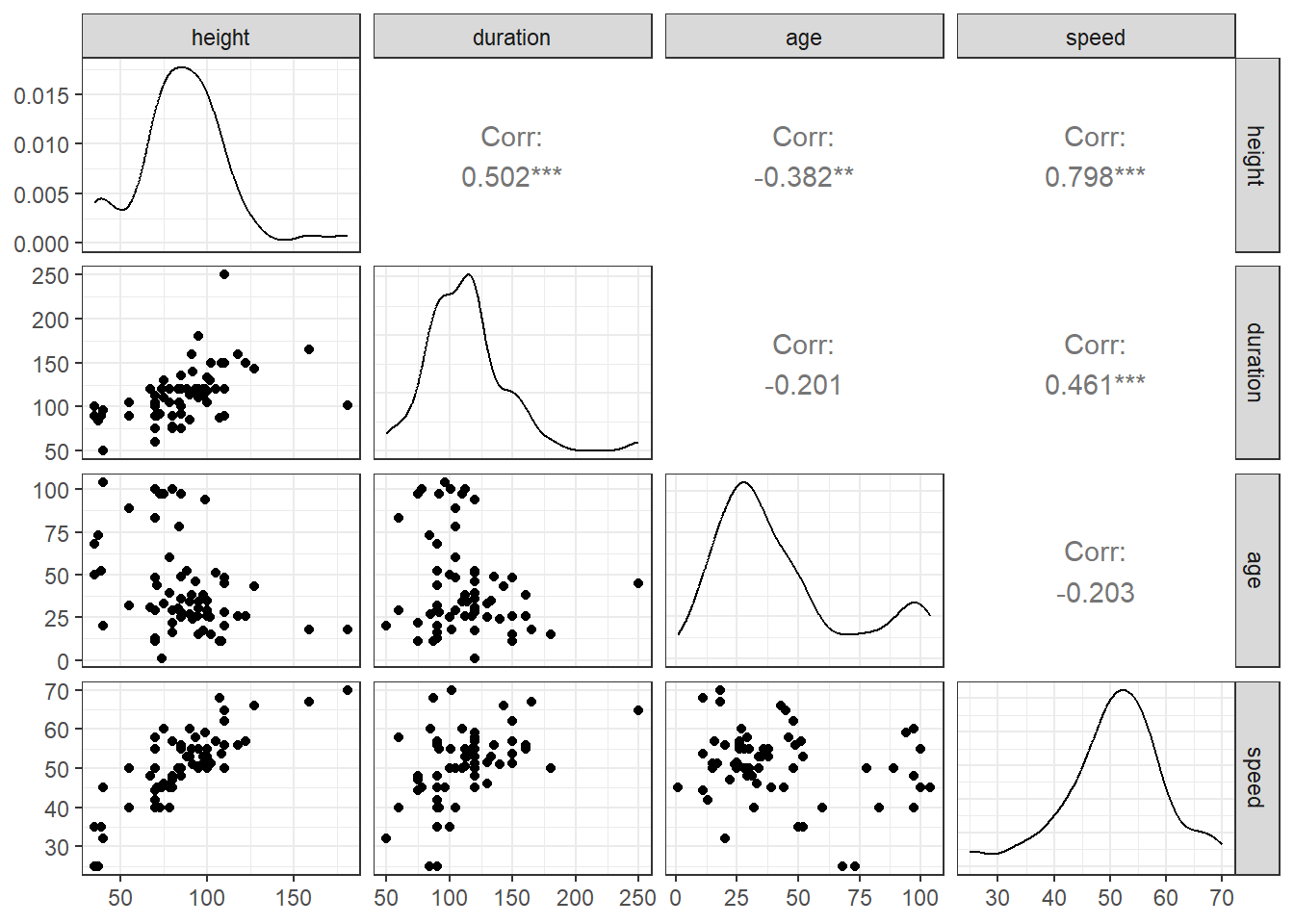
11.5 Scatterplot Matrix
We’ll use the ggpairs function from the GGally package when I want to build a combined matrix of scatterplots and correlation coefficients, as displayed below:
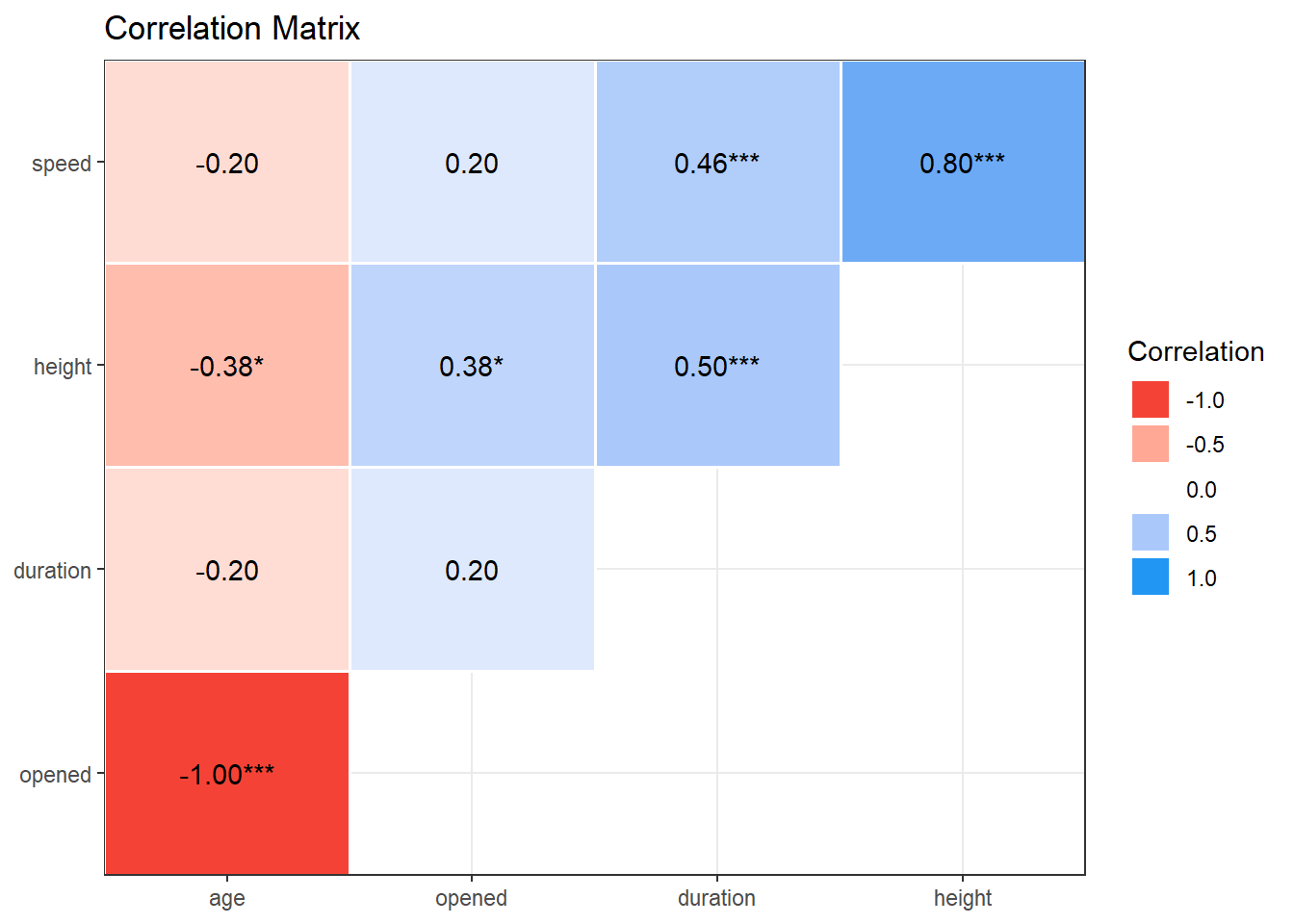
11.6 Correlation Matrix
Behold the complete set of correlation coefficients using the Pearson’s approach for the complete cases in the coasters data.
res_c <- correlation(coast_cc)
res_c# Correlation Matrix (pearson-method)
Parameter1 | Parameter2 | r | 95% CI | t(62) | p
--------------------------------------------------------------------
speed | height | 0.80 | [ 0.69, 0.87] | 10.43 | < .001***
speed | duration | 0.46 | [ 0.24, 0.63] | 4.09 | < .001***
speed | opened | 0.20 | [-0.04, 0.43] | 1.63 | 0.429
speed | age | -0.20 | [-0.43, 0.04] | -1.63 | 0.429
height | duration | 0.50 | [ 0.29, 0.67] | 4.58 | < .001***
height | opened | 0.38 | [ 0.15, 0.57] | 3.26 | 0.011*
height | age | -0.38 | [-0.57, -0.15] | -3.26 | 0.011*
duration | opened | 0.20 | [-0.05, 0.43] | 1.62 | 0.429
duration | age | -0.20 | [-0.43, 0.05] | -1.62 | 0.429
opened | age | -1.00 | [-1.00, -1.00] | -Inf | < .001***
p-value adjustment method: Holm (1979)
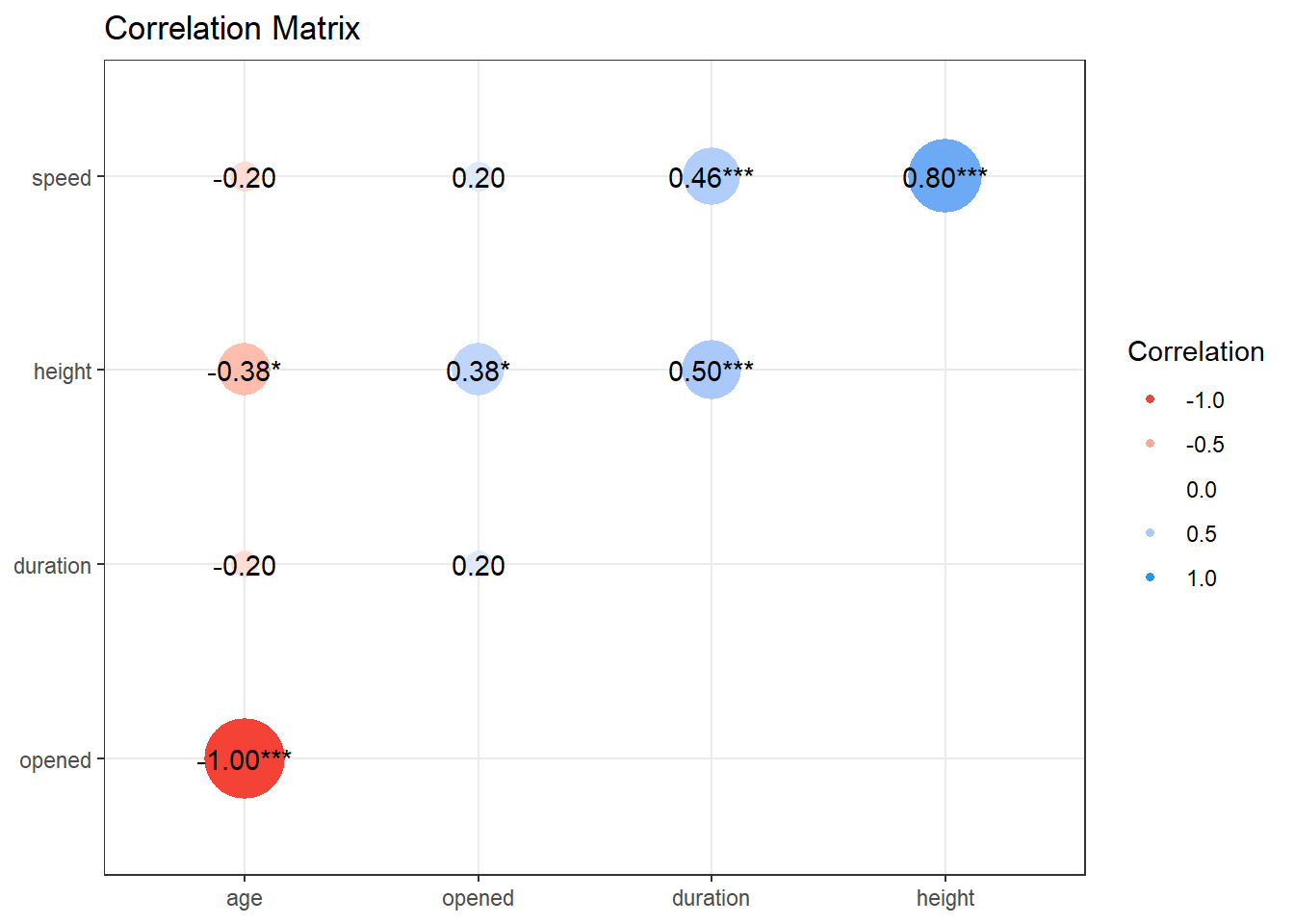
Observations: 64Once we have these results, we can plot them in several ways, in addition to the scatterplot matrix we’ve seen. These include:
and
11.6.1 Using a different measure
We can also obtain a correlation matrix using Spearman’s rank correlation, as follows:
res8 <- correlation(coast_cc, method = "spearman")
res8# Correlation Matrix (spearman-method)
Parameter1 | Parameter2 | rho | 95% CI | S | p
-----------------------------------------------------------------------
speed | height | 0.72 | [ 0.57, 0.82] | 12371.84 | < .001***
speed | duration | 0.45 | [ 0.23, 0.63] | 23898.81 | 0.001**
speed | opened | 0.17 | [-0.09, 0.40] | 36461.32 | 0.768
speed | age | -0.17 | [-0.40, 0.09] | 50898.68 | 0.768
height | duration | 0.60 | [ 0.41, 0.74] | 17308.98 | < .001***
height | opened | 0.39 | [ 0.15, 0.58] | 26851.92 | 0.010*
height | age | -0.39 | [-0.58, -0.15] | 60508.08 | 0.010*
duration | opened | 0.13 | [-0.12, 0.37] | 37811.21 | 0.768
duration | age | -0.13 | [-0.37, 0.12] | 49548.79 | 0.768
opened | age | -1.00 | [-1.00, -1.00] | 87360.00 | < .001***
p-value adjustment method: Holm (1979)
Observations: 6411.7 Modeling Speed with Height
Let’s start with a linear model (fit using ordinary least squares) to predict speed using height across our 64 coasters.
11.7.1 Fitting the Model
fit1 <- lm(speed ~ height, data = coast_cc)11.7.2 Parameter Estimates
model_parameters(fit1, ci = 0.95) |> kable(digits = 2)| Parameter | Coefficient | SE | CI | CI_low | CI_high | t | df_error | p |
|---|---|---|---|---|---|---|---|---|
| (Intercept) | 26.95 | 2.38 | 0.95 | 22.18 | 31.71 | 11.31 | 62 | 0 |
| height | 0.28 | 0.03 | 0.95 | 0.22 | 0.33 | 10.43 | 62 | 0 |
Good ways to interpret these coefficients include:
- When comparing any two coasters whose heights are one foot apart, we expect a speed that is, on average, 0.28 miles per hour faster, for the taller coaster.
- Under the fitted model, the average difference in coaster speed between two coasters whose height differs by ten feet is 2.8 MPH.
Note that we multiplied the height difference by ten, so we needed to multiply the average difference by 10, as well, in our second response.
11.7.3 Performance of the Model
How well does our model fit1 perform? Some key summaries (applicable to any of the linear models we’ve fit with ordinary least squares so far in this book, too) are shown below.
model_performance(fit1)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
---------------------------------------------------------------
403.859 | 404.259 | 410.336 | 0.637 | 0.631 | 5.416 | 5.503While we’ll focus on just a few of these measures in this chapter, specifically R2 and RMSE, here is a description of all of the summaries.
- AIC: Akaike’s Information Criterion
- AICc: Second-order (or small sample) AIC with a correction for small sample sizes
- BIC: Bayesian Information Criterion
AIC, AICc and BIC are used when comparing one or more models for the same outcome. When comparing models fitted by maximum likelihood (like ordinary least squares linear models), the smaller the AIC or BIC, the better the fit. See the R documentation for these information criteria here.
- R2: r-squared value
- R2_adj: adjusted r-squared
The R-squared (\(R^2\)) measure, also called the coefficient of determination, describes how much of the variation in our outcome can be explained using our model (and its predictors.) \(R^2\) falls between 0 and 1, and the closer it is to 1, the better the model fits our data. In a simple linear regression model such as we fit here (with one predictor), the \(R^2\) value is also the square of the Pearson correlation coefficient. Note also that in discussing ANOVA previously, we called the \(R^2\) value \(\eta^2\).
An adjusted R-squared measure, is an index (so it’s no longer a proportion of anything) used to compare different models (usually using different sets of predictors) that are fit to predict the same outcome. While adjusted \(R^2\) usually falls between 0 and 1, it can also be negative, and its formula takes into account both the number of observations available and the number of predictors in the model. The idea is to reduce the temptation to overfit the data, by penalizing the \(R^2\) value a little for each predictor. The adjusted \(R^2\) measure is always no larger than \(R^2\). See the documentation for these measures in the performance package (part of easystats.)
- RMSE: root mean squared error
- SIGMA: residual standard deviation, see insight::get_sigma()
The RMSE is the square root of the variance of the residuals and indicates the absolute fit of the model to the data (difference between observed data to model’s predicted values). It can be interpreted as the standard deviation of the unexplained variance, and is in the same units as the response variable. Lower values indicate better model fit. (Source).
Linear models assume that their residuals are drawn from a Normal distribution with mean 0 and standard deviation equal to sigma (\(\sigma\)). The residual standard deviation sigma indicates the accuracy for a model to predict our outcome. \(\sigma\) also indicates that the predicted outcome will be within \(\pm 1 \sigma\) units of the observed outcome for approximately 68% of the data points, for example. See the discussion of sigma here.
11.8 Checking a Linear Model
The main assumptions of any linear model are:
- linearity: we assume that the outcome is linearly related to our predictor
- constant variance (homoscedasticity): we assume that the variation of our outcome is about the same regardless of the value of our predictor
- normal distribution: we assume that the errors around the regression model at any specified values of the x-variables follow an approximately Normal distribution.
11.8.1 Posterior Predictive Checks
Next, we’ll walk through what each of these plots is doing, one at a time.
diagnostic_plots <- plot(check_model(fit1, panel = FALSE))For confidence bands, please install `qqplotr`.diagnostic_plots[[1]]
11.8.2 Checking Linearity
diagnostic_plots[[2]]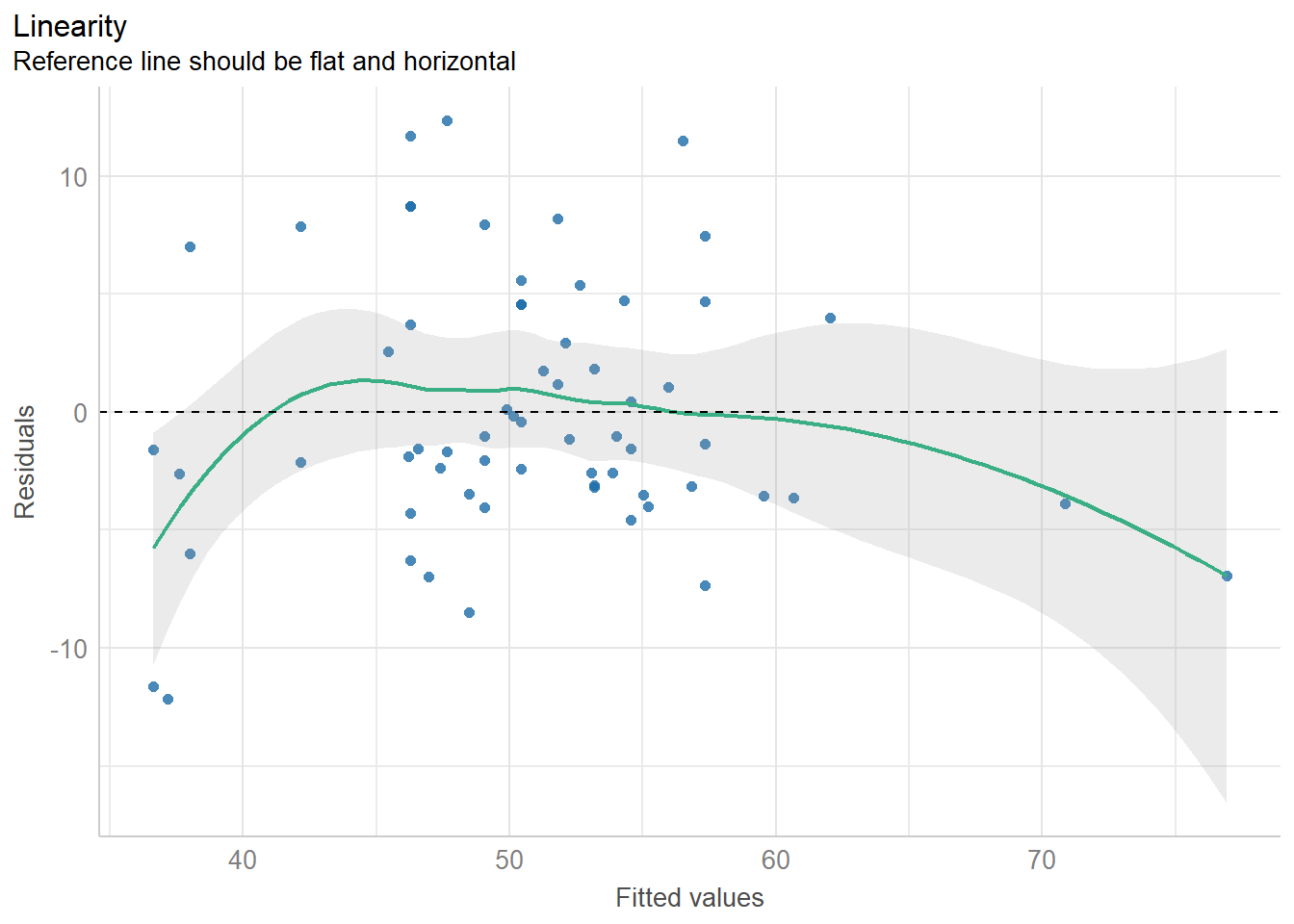
11.8.3 Checking for Homogeneity of Variance
diagnostic_plots[[3]]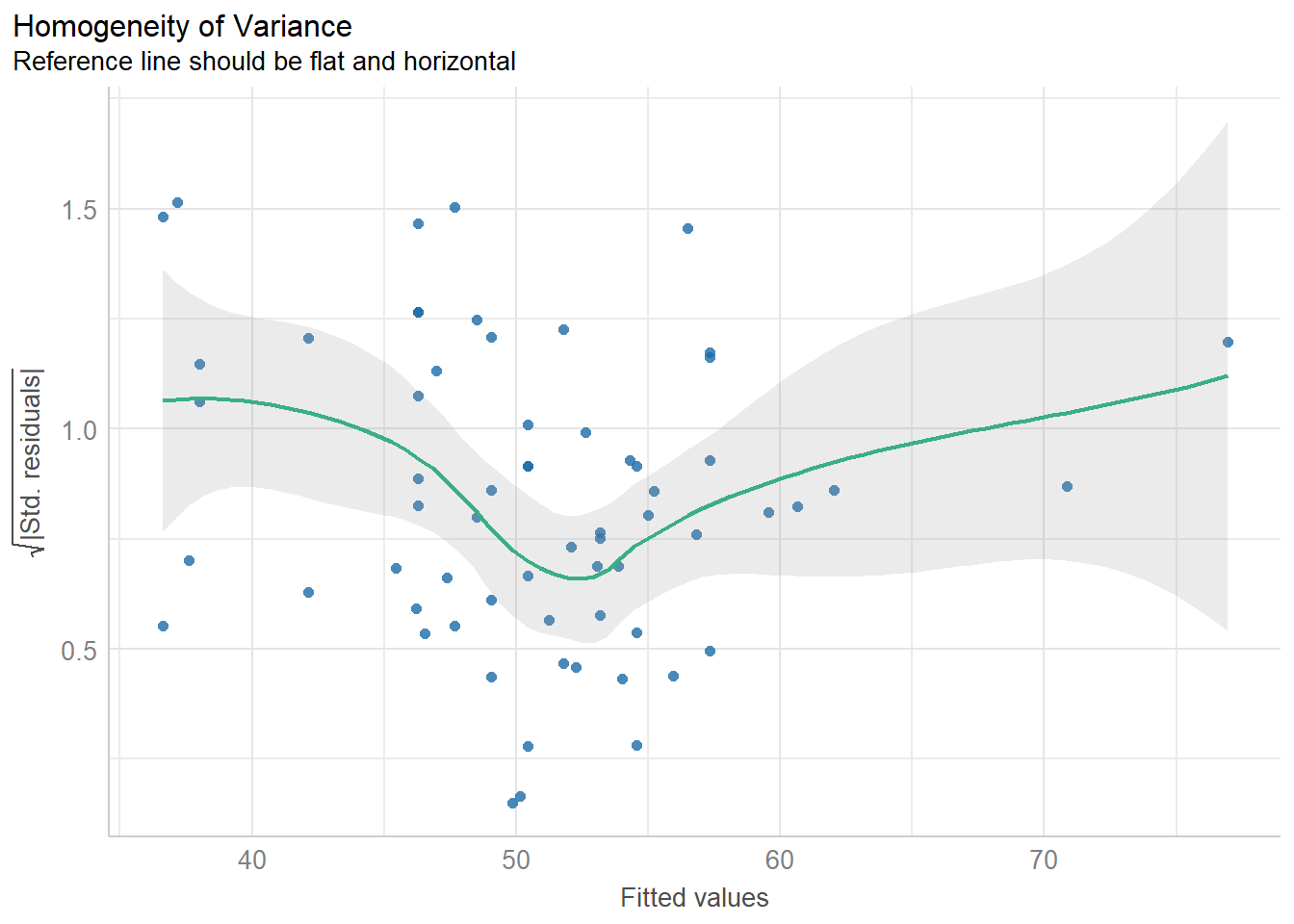
11.8.4 Checking for Influential Observations
diagnostic_plots[[4]]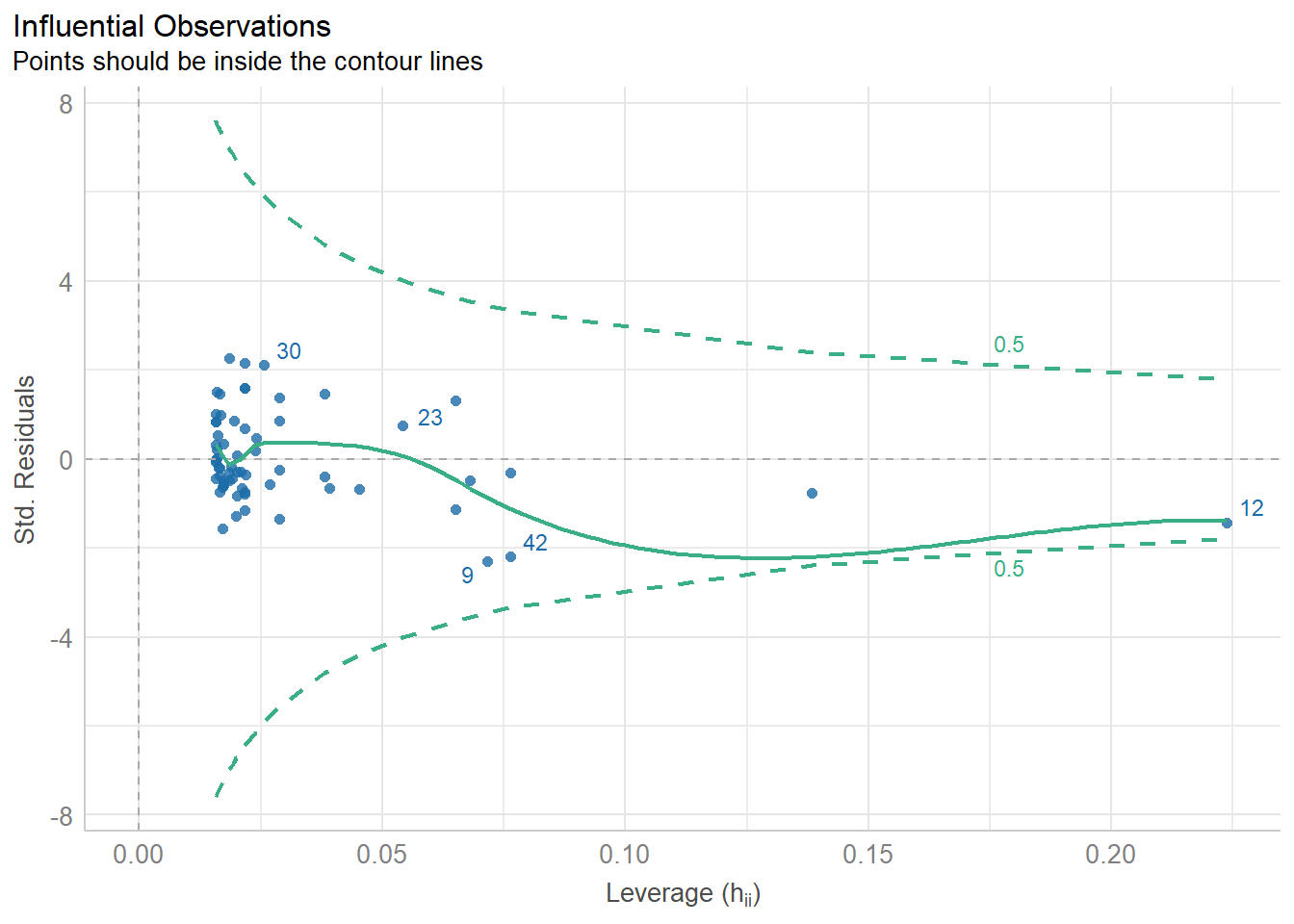
11.8.5 Checking for Normality of Residuals
diagnostic_plots[[5]]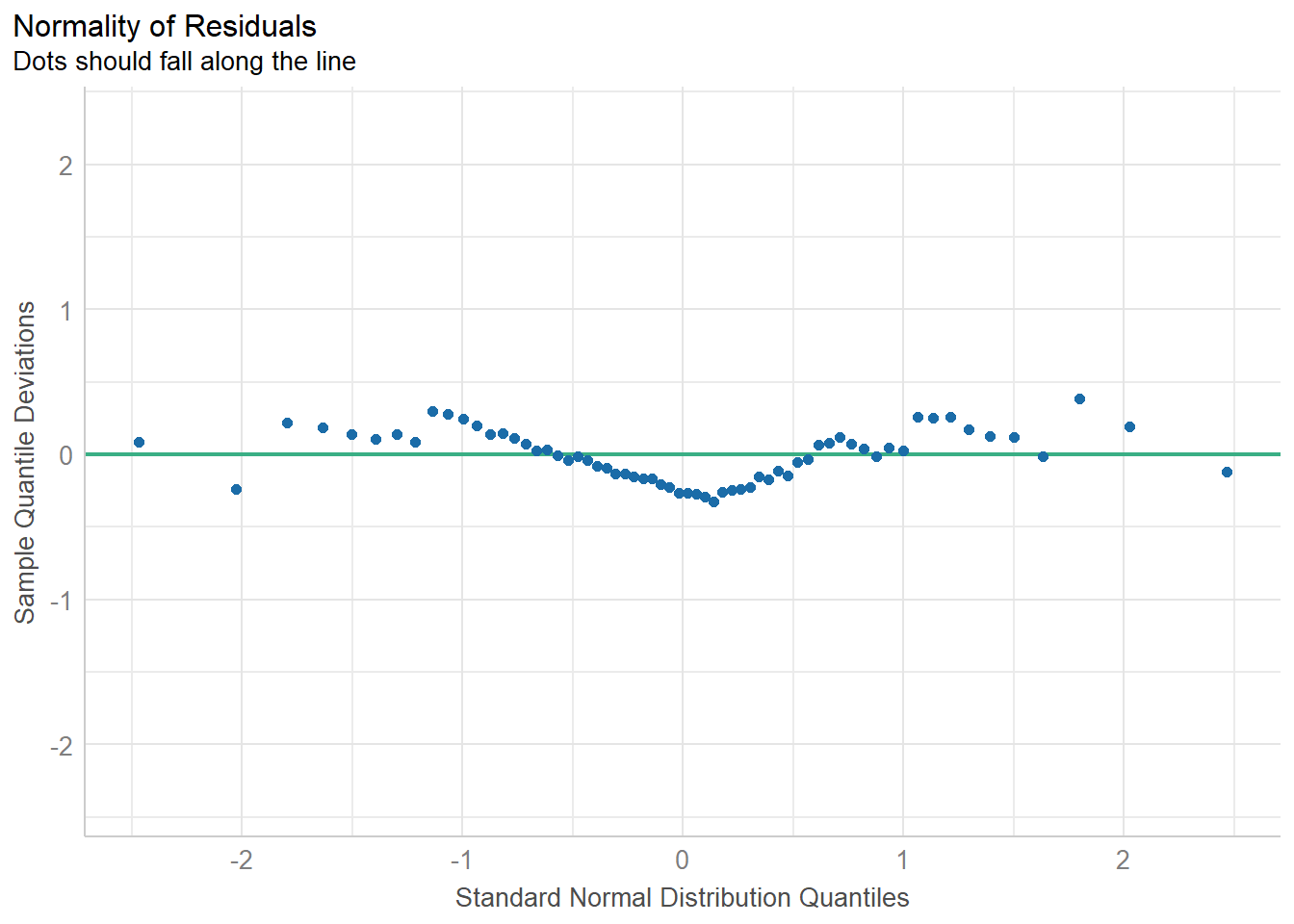
11.8.6 Running All of the Checks
You will want to include
#| fig-height: 9
at the start of the R code chunk where you run check_model() in order to give the plots some vertical space to breathe.
check_model(fit1)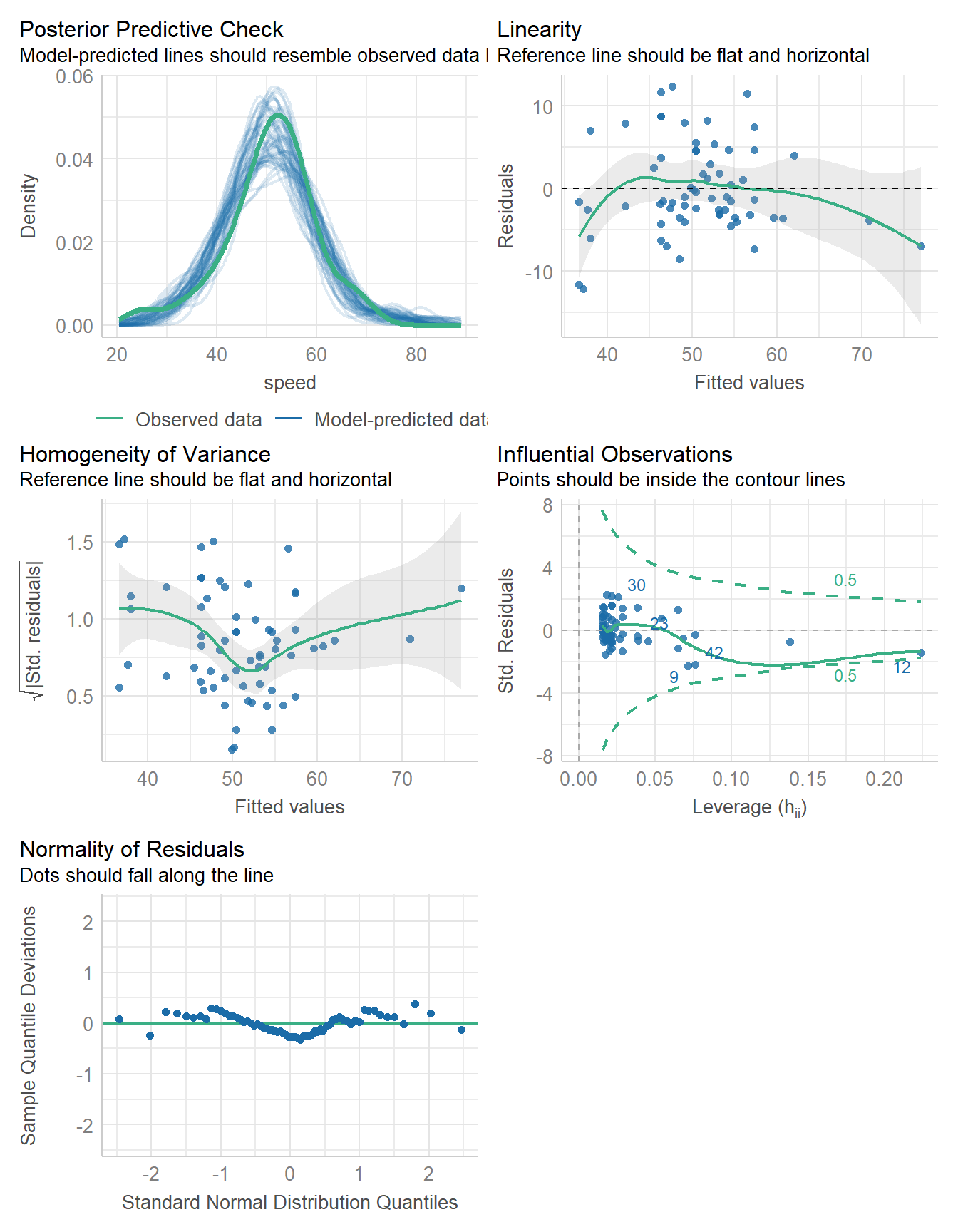
- See the check_model() web page at easystats (performance) for more details.
check_normality(fit1)OK: residuals appear as normally distributed (p = 0.104).check_heteroskedasticity(fit1)OK: Error variance appears to be homoscedastic (p = 0.077).11.9 Bayesian linear model for Speed with Height
11.9.1 Fitting the model
Here, we fit the Bayesian model, assuming weakly informative priors on the coefficients.
11.9.2 Parameter Estimates
model_parameters(fit2, ci = 0.95) |> kable(digits = 2)| Parameter | Median | CI | CI_low | CI_high | pd | Rhat | ESS | Prior_Distribution | Prior_Location | Prior_Scale |
|---|---|---|---|---|---|---|---|---|---|---|
| (Intercept) | 26.90 | 0.95 | 22.05 | 31.97 | 1 | 1 | 3770.92 | normal | 50.76 | 22.65 |
| height | 0.28 | 0.95 | 0.22 | 0.33 | 1 | 1 | 3516.32 | normal | 0.00 | 0.87 |
11.9.3 Performance of the Model
model_performance(fit2)# Indices of model performance
ELPD | ELPD_SE | LOOIC | LOOIC_SE | WAIC | R2 | R2 (adj.) | RMSE | Sigma
-------------------------------------------------------------------------------------
-202.245 | 5.520 | 404.491 | 11.039 | 404.407 | 0.630 | 0.614 | 5.416 | 5.55211.9.4 Checking the Model
check_model(fit2)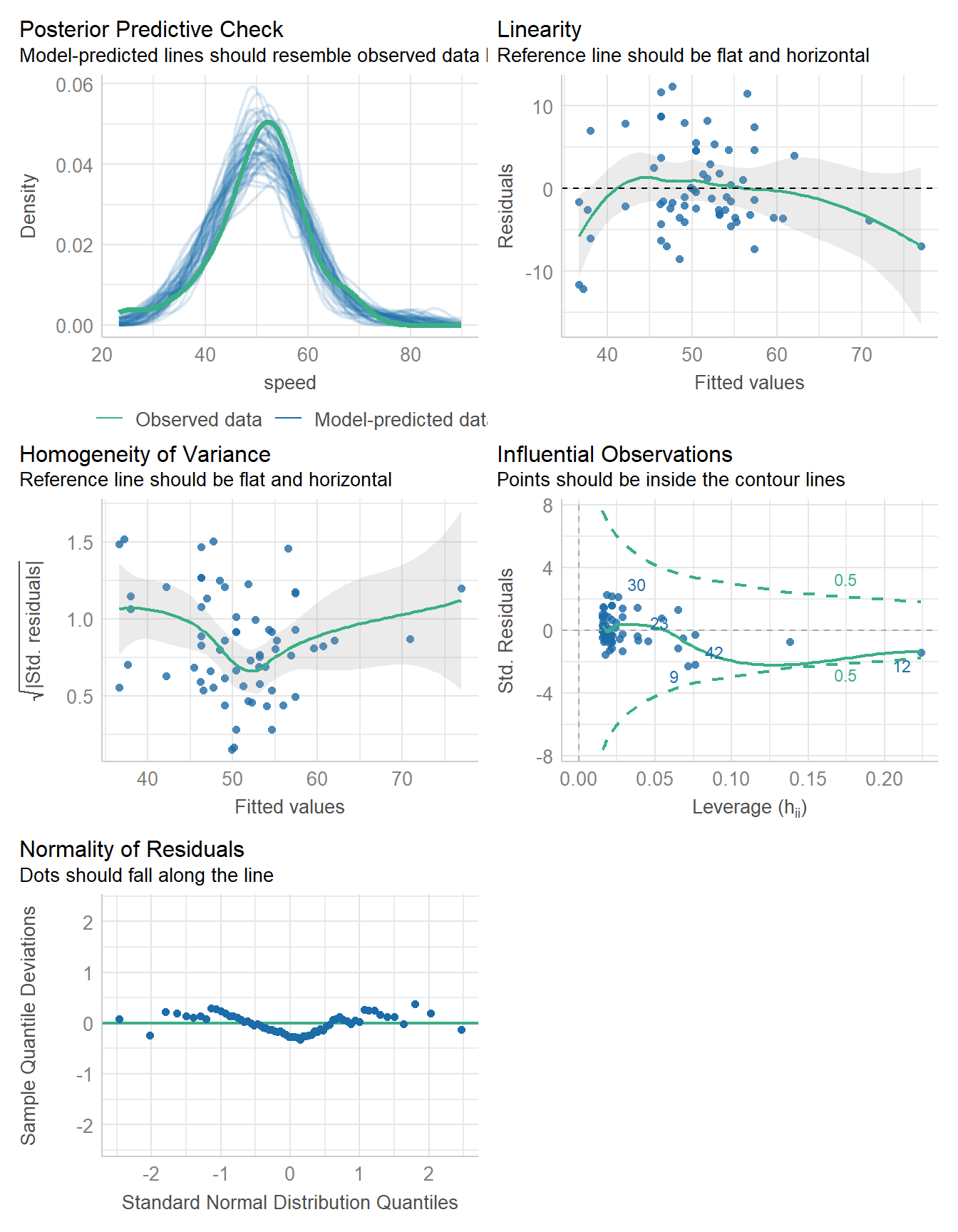
11.10 Predicting Speed with Duration
Next, consider the relationship between duration and speed.
ggplot(coast_cc, aes(x = duration, y = speed)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x, col = "red") +
geom_smooth(method = "loess", formula = y ~ x, se = FALSE, col = "blue") +
labs(title = "Association of Speed with Duration") +
stat_regline_equation(label.x = 200, label.y = 40) +
stat_cor(aes(label = after_stat(rr.label)),
label.x = 200, label.y = 35)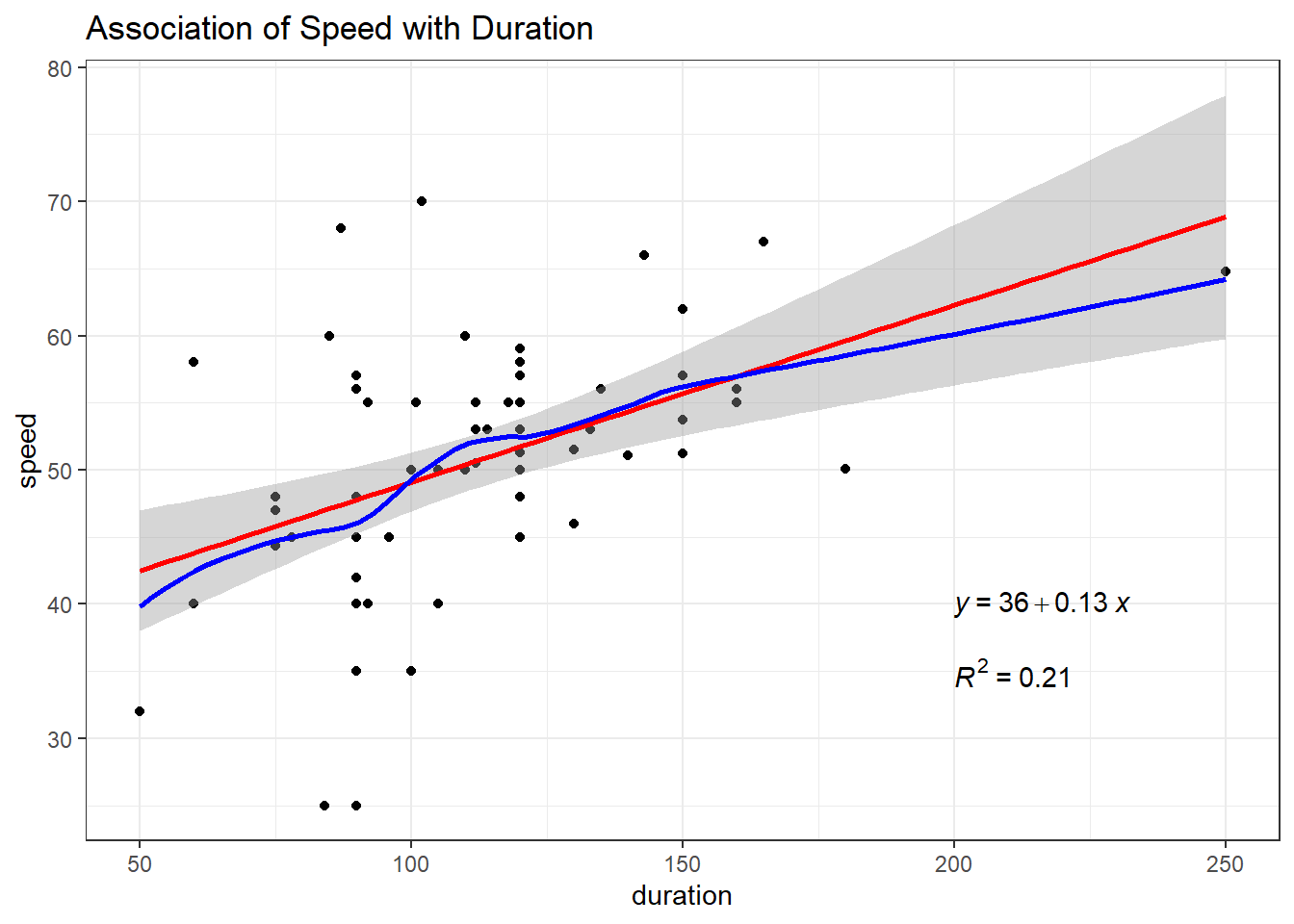
11.10.1 Ordinary Least Squares
fit3 <- lm(speed ~ duration, data = coast_cc)
model_parameters(fit3, ci = 0.95) |> kable(digits = 2)| Parameter | Coefficient | SE | CI | CI_low | CI_high | t | df_error | p |
|---|---|---|---|---|---|---|---|---|
| (Intercept) | 35.91 | 3.77 | 0.95 | 28.38 | 43.44 | 9.53 | 62 | 0 |
| duration | 0.13 | 0.03 | 0.95 | 0.07 | 0.20 | 4.09 | 62 | 0 |
model_performance(fit3) |> kable()| AIC | AICc | BIC | R2 | R2_adjusted | RMSE | Sigma |
|---|---|---|---|---|---|---|
| 453.4355 | 453.8355 | 459.9122 | 0.2126508 | 0.1999516 | 7.97771 | 8.105361 |
check_model(fit3)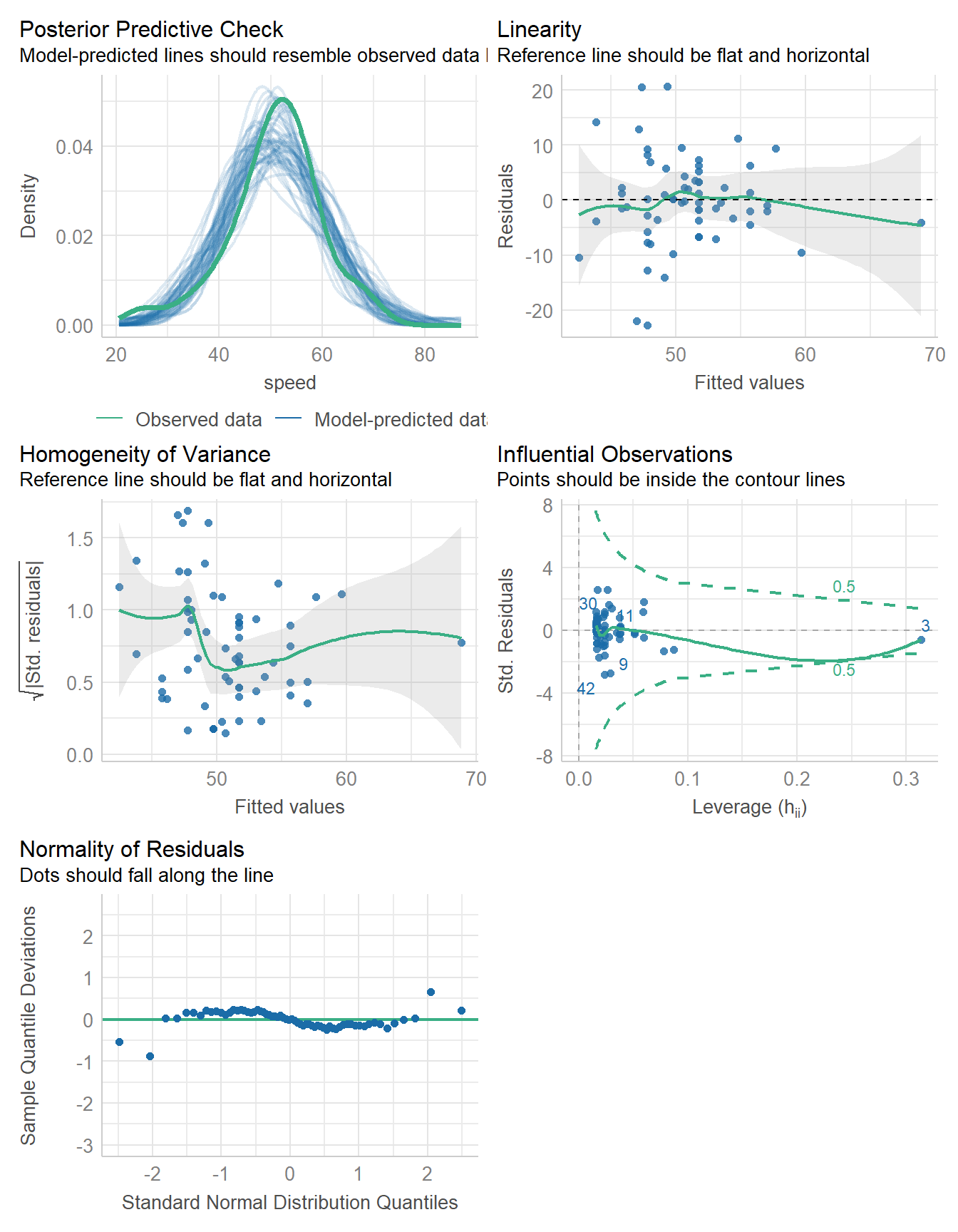
11.10.2 Bayesian linear model fit
set.seed(431)
fit4 <- stan_glm(speed ~ duration, data = coast_cc, refresh = 0)
model_parameters(fit4, ci = 0.95) |> kable(digits = 2)| Parameter | Median | CI | CI_low | CI_high | pd | Rhat | ESS | Prior_Distribution | Prior_Location | Prior_Scale |
|---|---|---|---|---|---|---|---|---|---|---|
| (Intercept) | 35.95 | 0.95 | 28.34 | 43.33 | 1 | 1 | 3722.28 | normal | 50.76 | 22.65 |
| duration | 0.13 | 0.95 | 0.07 | 0.20 | 1 | 1 | 3704.16 | normal | 0.00 | 0.71 |
model_performance(fit4)# Indices of model performance
ELPD | ELPD_SE | LOOIC | LOOIC_SE | WAIC | R2 | R2 (adj.) | RMSE | Sigma
-------------------------------------------------------------------------------------
-227.090 | 7.486 | 454.181 | 14.972 | 454.127 | 0.209 | 0.180 | 7.978 | 8.168check_model(fit4)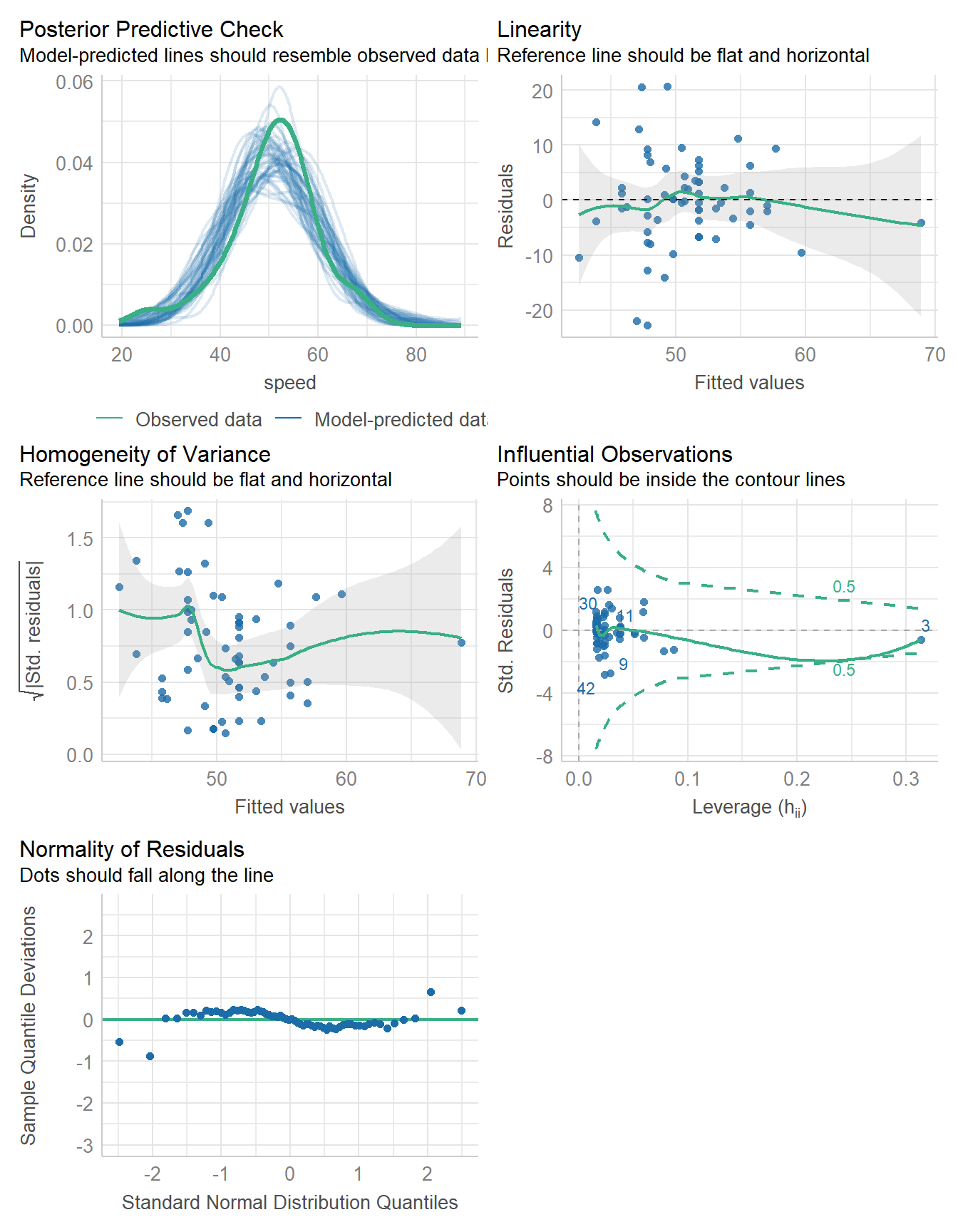
11.11 Predicting Speed with Age
Finally, are newer coasters faster? Consider the relationship between age and speed.
ggplot(coast_cc, aes(x = age, y = speed)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x, col = "red") +
geom_smooth(method = "loess", formula = y ~ x, se = FALSE, col = "blue") +
labs(title = "Association of Speed with Age") +
stat_regline_equation(label.x = 65, label.y = 65) +
stat_cor(aes(label = after_stat(rr.label)),
label.x = 65, label.y = 60)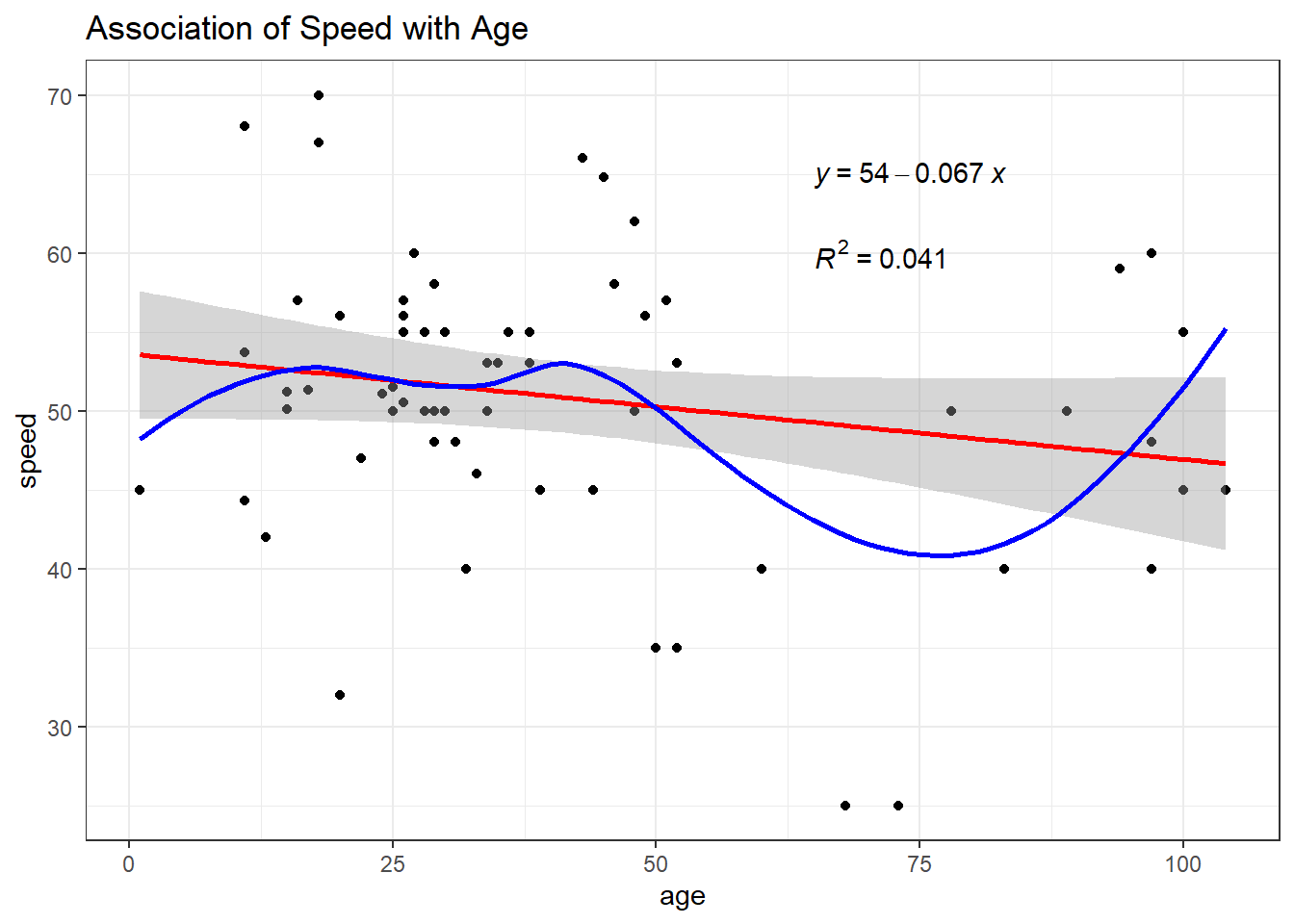
11.11.1 Ordinary Least Squares Fit
| Parameter | Coefficient | SE | CI | CI_low | CI_high | t | df_error | p |
|---|---|---|---|---|---|---|---|---|
| (Intercept) | 53.61 | 2.07 | 0.95 | 49.47 | 57.75 | 25.86 | 62 | 0.00 |
| age | -0.07 | 0.04 | 0.95 | -0.15 | 0.01 | -1.63 | 62 | 0.11 |
model_performance(fit5)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
---------------------------------------------------------------
466.039 | 466.439 | 472.516 | 0.041 | 0.026 | 8.803 | 8.944check_model(fit5)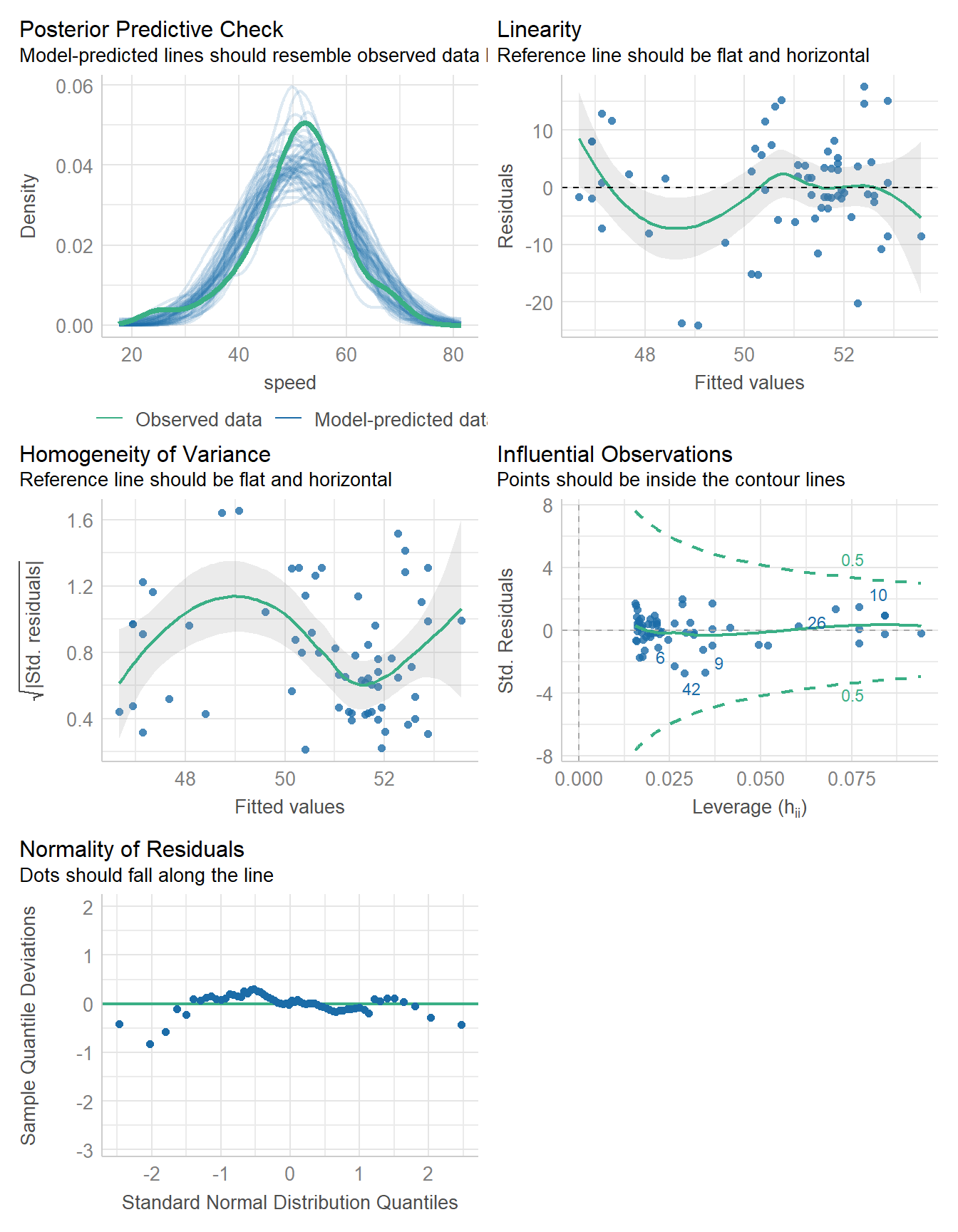
11.11.2 Bayesian linear model fit
set.seed(431)
fit6 <- stan_glm(speed ~ age, data = coast_cc, refresh = 0)
model_parameters(fit6, ci = 0.95) |> kable(digits = 2)| Parameter | Median | CI | CI_low | CI_high | pd | Rhat | ESS | Prior_Distribution | Prior_Location | Prior_Scale |
|---|---|---|---|---|---|---|---|---|---|---|
| (Intercept) | 53.54 | 0.95 | 49.57 | 57.64 | 1.00 | 1 | 3569.63 | normal | 50.76 | 22.65 |
| age | -0.07 | 0.95 | -0.15 | 0.01 | 0.95 | 1 | 3753.96 | normal | 0.00 | 0.82 |
model_performance(fit6)# Indices of model performance
ELPD | ELPD_SE | LOOIC | LOOIC_SE | WAIC | R2 | R2 (adj.) | RMSE | Sigma
-------------------------------------------------------------------------------------
-233.199 | 6.670 | 466.398 | 13.341 | 466.367 | 0.039 | -0.003 | 8.803 | 8.971check_model(fit6)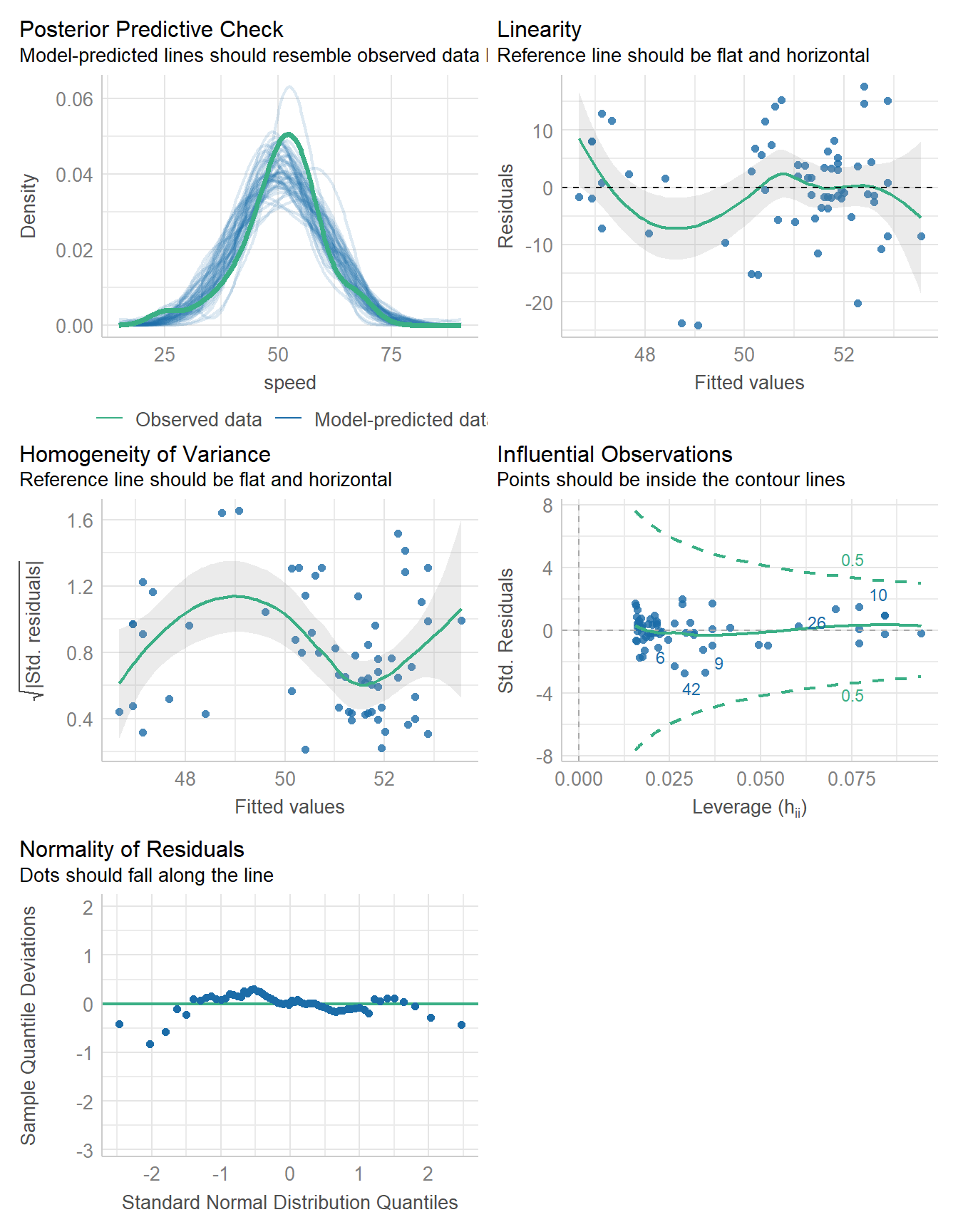
11.12 For More Information
- Chapter 7 of Çetinkaya-Rundel and Hardin (2024) is about linear regression with a single predictor.
- This tutorial from the
bayestestRpackage (part ofeasystats) uses simple linear regression with bothlm()and Bayesian approaches. - Here is a nice tutorial using R on Simple Linear Regression that addresses some of this chapter’s issues.
- Wikipedia’s page on Linear regression provides lots of useful details.
- Plotting Functions for the correlation package
- Correlation Types
My favorite amusement park is Kennywood Park, just outside of Pittsburgh.↩︎