18 Multiple Regression with Two Predictors
18.1 R setup for this chapter
Appendix A lists all R packages used in this book, and also provides R session information. Appendix B describes the 431-Love.R script, and demonstrates its use.
18.2 Child Care Costs from Tidy Tuesday
Appendix C provides further guidance on pulling data from other systems into R, while Appendix D gives more information (including download links) for all data sets used in this book.
The data for this example is a subset of data described at the Tidy Tuesday repository for 2023-05-09, on Child Care Costs.
The data source is the National Database of Childcare Prices.
The National Database of Childcare Prices (NDCP) is the most comprehensive federal source of childcare prices at the county level. The database offers childcare price data by childcare provider type, age of children, and county characteristics. Data are available from 2008 to 2018.
We will focus on the 2018 data at the county level for the US, and we will read in the data using the tidytuesdayR package.
The data we’re interested in comes in two separate files, one called childcare_costs, and the other called counties.
tuesdata <- tidytuesdayR::tt_load("2023-05-09")---- Compiling #TidyTuesday Information for 2023-05-09 ----
--- There are 2 files available ---
── Downloading files ───────────────────────────────────────────────────────────
1 of 2: "childcare_costs.csv"
2 of 2: "counties.csv"childcare_costs <- tuesdata$childcare_costs
counties <- tuesdata$countieshead(childcare_costs)# A tibble: 6 × 61
county_fips_code study_year unr_16 funr_16 munr_16 unr_20to64 funr_20to64
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1001 2008 5.42 4.41 6.32 4.6 3.5
2 1001 2009 5.93 5.72 6.11 4.8 4.6
3 1001 2010 6.21 5.57 6.78 5.1 4.6
4 1001 2011 7.55 8.13 7.03 6.2 6.3
5 1001 2012 8.6 8.88 8.29 6.7 6.4
6 1001 2013 9.39 10.3 8.56 7.3 7.6
# ℹ 54 more variables: munr_20to64 <dbl>, flfpr_20to64 <dbl>,
# flfpr_20to64_under6 <dbl>, flfpr_20to64_6to17 <dbl>,
# flfpr_20to64_under6_6to17 <dbl>, mlfpr_20to64 <dbl>, pr_f <dbl>,
# pr_p <dbl>, mhi_2018 <dbl>, me_2018 <dbl>, fme_2018 <dbl>, mme_2018 <dbl>,
# total_pop <dbl>, one_race <dbl>, one_race_w <dbl>, one_race_b <dbl>,
# one_race_i <dbl>, one_race_a <dbl>, one_race_h <dbl>, one_race_other <dbl>,
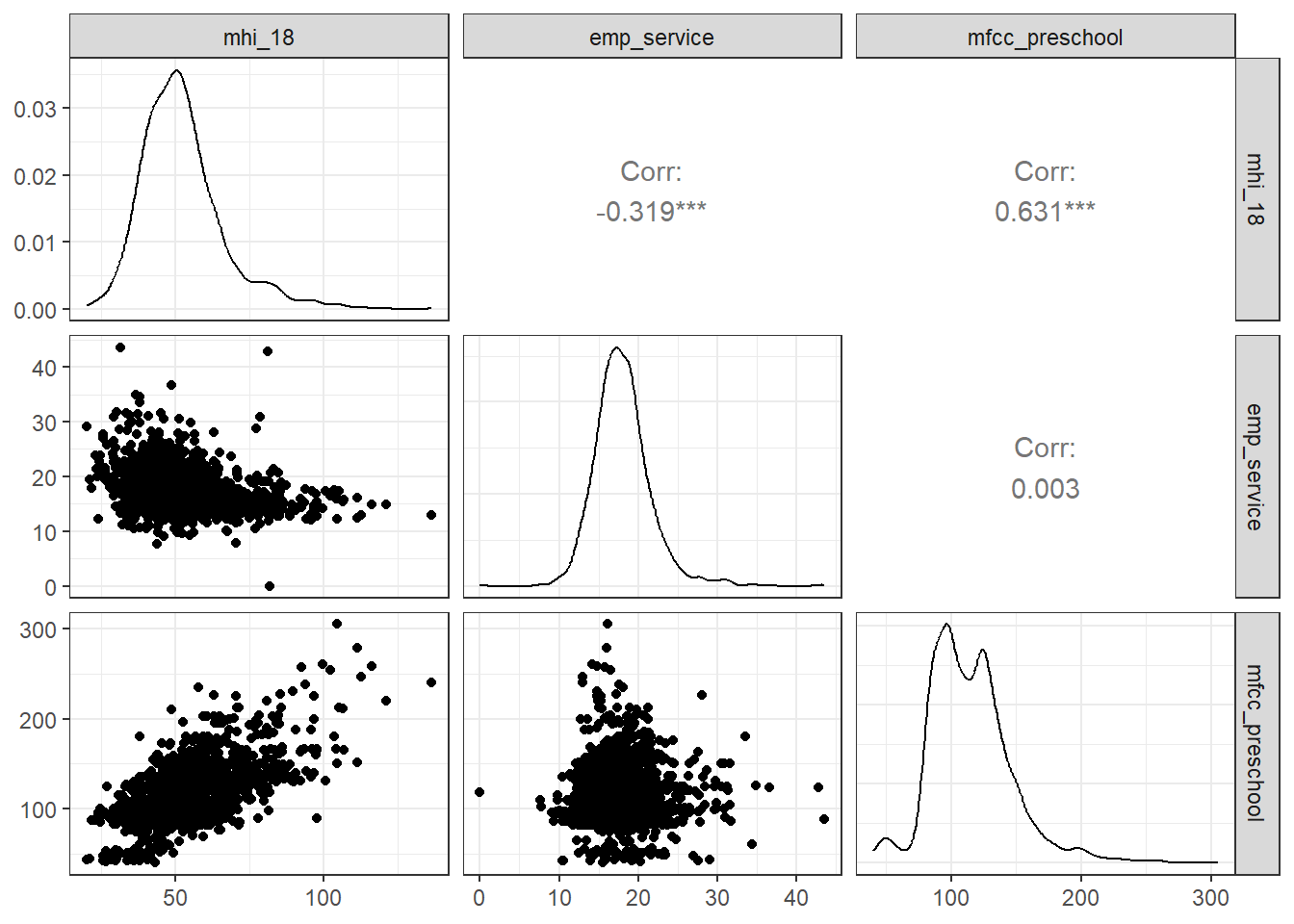
# two_races <dbl>, hispanic <dbl>, households <dbl>, …First, we work with the childcare_costs data to select the study_year of 2018, then collect the following variables (besides the county_fips_code we’ll use to link to the counties data and the study_year):
- Our outcome will be
mfcc_preschool(Aggregated weekly, full-time median price charged for Family Childcare for preschoolers (i.e. aged 36 through 54 months).- Actually, we’ll use the square root of
mfcc_preschool, which I’ll callsqrt_priceas our main outcome, which I decided to do after exploring the data a bit.
- Actually, we’ll use the square root of
- Our first predictor is
mhi_2018(Median household income expressed in 2018 dollars.) but recast asmhi_18(mhi_2018 / 1000) to express income in thousands of 2018 dollars, - Our other predictor is
emp_service(Percent of civilians employed in service occupations aged 16 years old and older in the county.)
In terms of dealing with missingness, we will drop all counties with missing values of the outcome, which is the only missing element .
costs <- childcare_costs |>
filter(study_year == 2018) |>
mutate(mhi_18 = mhi_2018/1000) |>
mutate(sqrt_price = sqrt(mfcc_preschool)) |>
select(county_fips_code, mfcc_preschool,
sqrt_price, mhi_18, emp_service) |>
filter(complete.cases(mfcc_preschool))
prop_miss_case(costs)[1] 0Next, we use left_join() to merge the childcare costs and the county information by the county_fips_code.
care_costs <- left_join(costs, counties, by = "county_fips_code") |>
rename(fips = county_fips_code,
county = county_name,
state = state_abbreviation) |>
mutate(fips = as.character(fips))
care_costs# A tibble: 2,348 × 8
fips mfcc_preschool sqrt_price mhi_18 emp_service county state_name state
<chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
1 1001 106. 10.3 58.8 15.9 Autauga … Alabama AL
2 1003 109. 10.4 56.0 18.1 Baldwin … Alabama AL
3 1005 77.1 8.78 34.2 14.6 Barbour … Alabama AL
4 1007 87.1 9.33 45.3 18.7 Bibb Cou… Alabama AL
5 1009 112. 10.6 48.7 13 Blount C… Alabama AL
6 1011 106. 10.3 32.2 17.1 Bullock … Alabama AL
7 1013 107. 10.3 39.1 16.0 Butler C… Alabama AL
8 1015 85.7 9.26 45.2 16.8 Calhoun … Alabama AL
9 1017 116. 10.8 39.9 16.0 Chambers… Alabama AL
10 1019 64.6 8.04 41.0 12.2 Cherokee… Alabama AL
# ℹ 2,338 more rows18.2.1 Partitioning the Data
If we like, we can then partition the data into separate training and testing samples. Here, we’ll put 70% of the rows (counties) in our training sample, and the remaining 30% into the test sample.
18.3 Fit model in training sample with lm
First, draw a picture with ggpairs from GGally of the training sample, so we can see the individual associations.
Next, we’ll fit the model, and obtain some basic summaries of the model’s parameters and its performance.
fit1 <- lm(sqrt_price ~ mhi_18 + emp_service, data = ccosts_train)model_parameters(fit1, ci = 0.95)Parameter | Coefficient | SE | 95% CI | t(1640) | p
----------------------------------------------------------------------
(Intercept) | 5.31 | 0.20 | [4.91, 5.71] | 26.04 | < .001
mhi 18 | 0.07 | 2.03e-03 | [0.07, 0.08] | 35.04 | < .001
emp service | 0.09 | 7.86e-03 | [0.07, 0.11] | 11.44 | < .001
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a Wald t-distribution approximation.model_performance(fit1)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
------------------------------------------------------------
4957.6 | 4957.6 | 4979.2 | 0.428 | 0.427 | 1.091 | 1.092Next, we’ll check a set of regression diagnostics to evaluate whether assumptions of linear regression appear to hold sufficiently well.
check_model(fit1)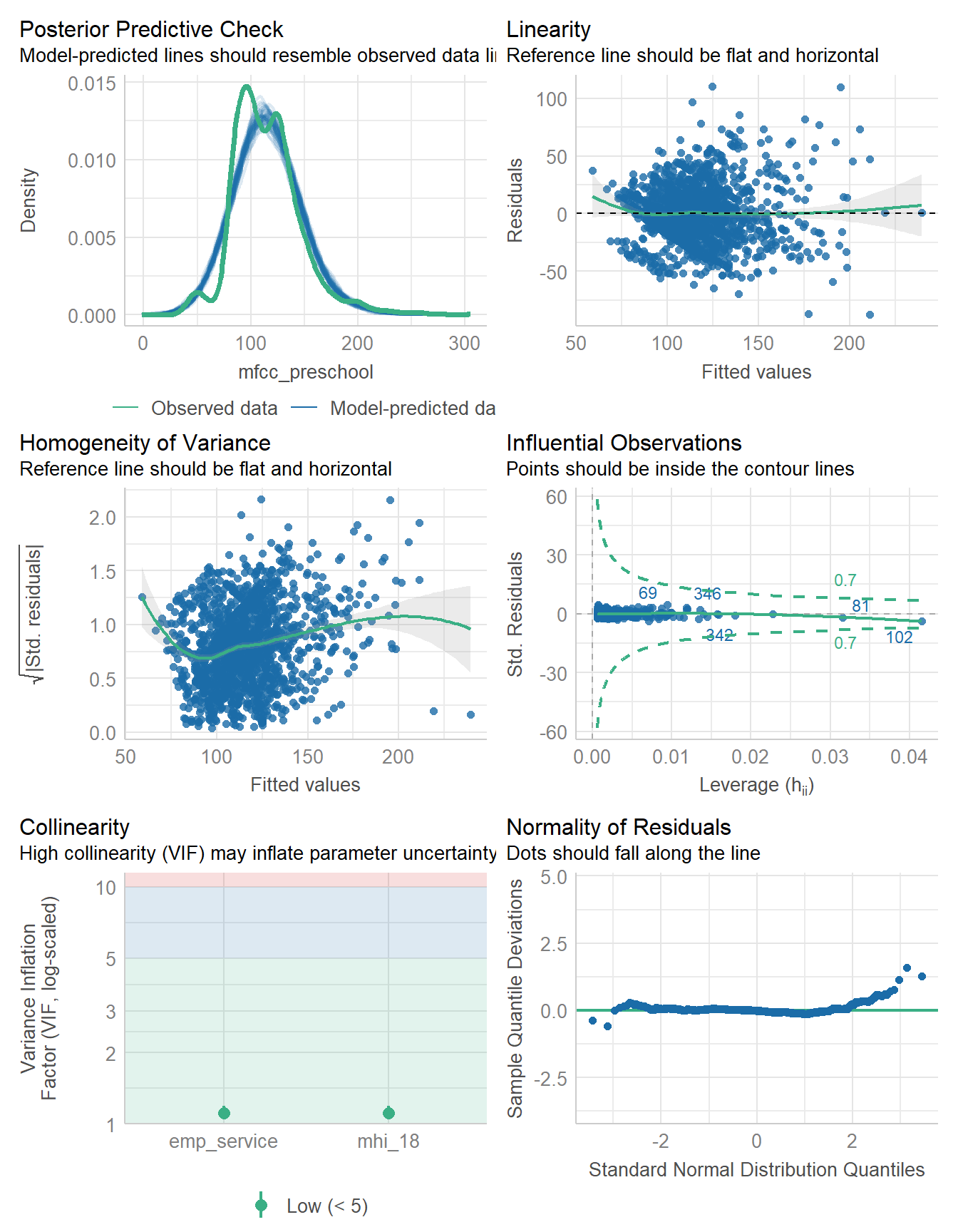
check_heteroscedasticity(fit1)Warning: Heteroscedasticity (non-constant error variance) detected (p < .001).check_normality(fit1)Warning: Non-normality of residuals detected (p < .001).18.4 Fitting an alternative model
Here, we’ll also consider fitting a model with only one predictor (the mhi_18) variable, to see whether the addition of emp_service leads to a meaningful improvement. I don’t expect this to be a better model than the model (fit1) that also includes emp_service in this case, but we’ll use this comparison to demonstrate an appealing approach to using training and testing samples.
fit2 <- lm(sqrt_price ~ mhi_18, data = ccosts_train)model_parameters(fit2, ci = 0.95)Parameter | Coefficient | SE | 95% CI | t(1641) | p
----------------------------------------------------------------------
(Intercept) | 7.32 | 0.11 | [7.11, 7.53] | 67.71 | < .001
mhi 18 | 0.06 | 2.00e-03 | [0.06, 0.07] | 31.88 | < .001
Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
using a Wald t-distribution approximation.model_performance(fit2)# Indices of model performance
AIC | AICc | BIC | R2 | R2 (adj.) | RMSE | Sigma
------------------------------------------------------------
5081.8 | 5081.8 | 5098.0 | 0.382 | 0.382 | 1.134 | 1.135check_model(fit2)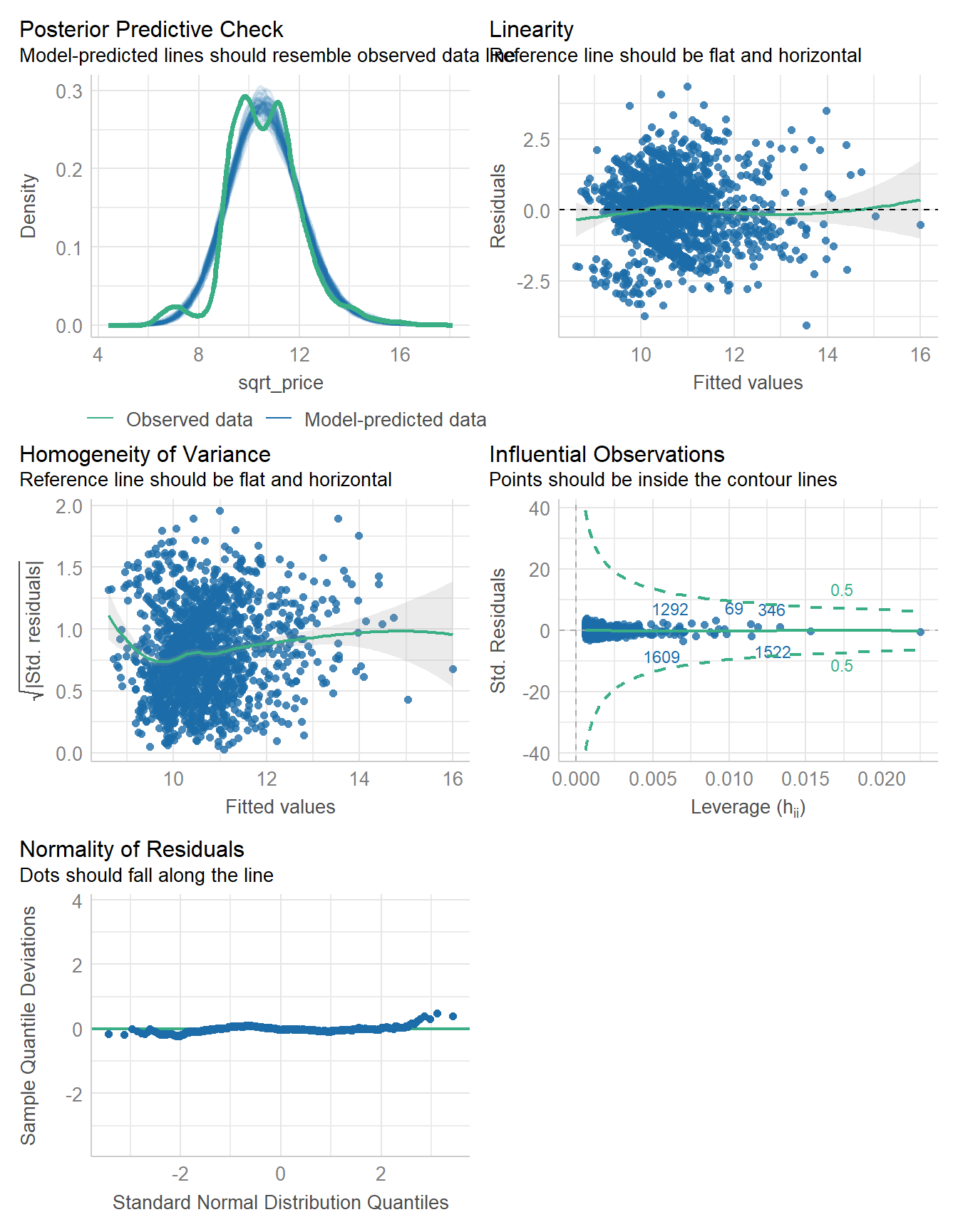
check_heteroscedasticity(fit2)Warning: Heteroscedasticity (non-constant error variance) detected (p < .001).check_normality(fit2)Warning: Non-normality of residuals detected (p < .001).18.5 Comparing the models (training sample)
We can compare the models within our training sample by comparing the performance statistics, as well as the quality of the diagnostic checks.
compare_performance(fit1, fit2)# Comparison of Model Performance Indices
Name | Model | AIC (weights) | AICc (weights) | BIC (weights) | R2
-----------------------------------------------------------------------
fit1 | lm | 4957.6 (>.999) | 4957.6 (>.999) | 4979.2 (>.999) | 0.428
fit2 | lm | 5081.8 (<.001) | 5081.8 (<.001) | 5098.0 (<.001) | 0.382
Name | R2 (adj.) | RMSE | Sigma
--------------------------------
fit1 | 0.427 | 1.091 | 1.092
fit2 | 0.382 | 1.134 | 1.135plot(compare_performance(fit1, fit2))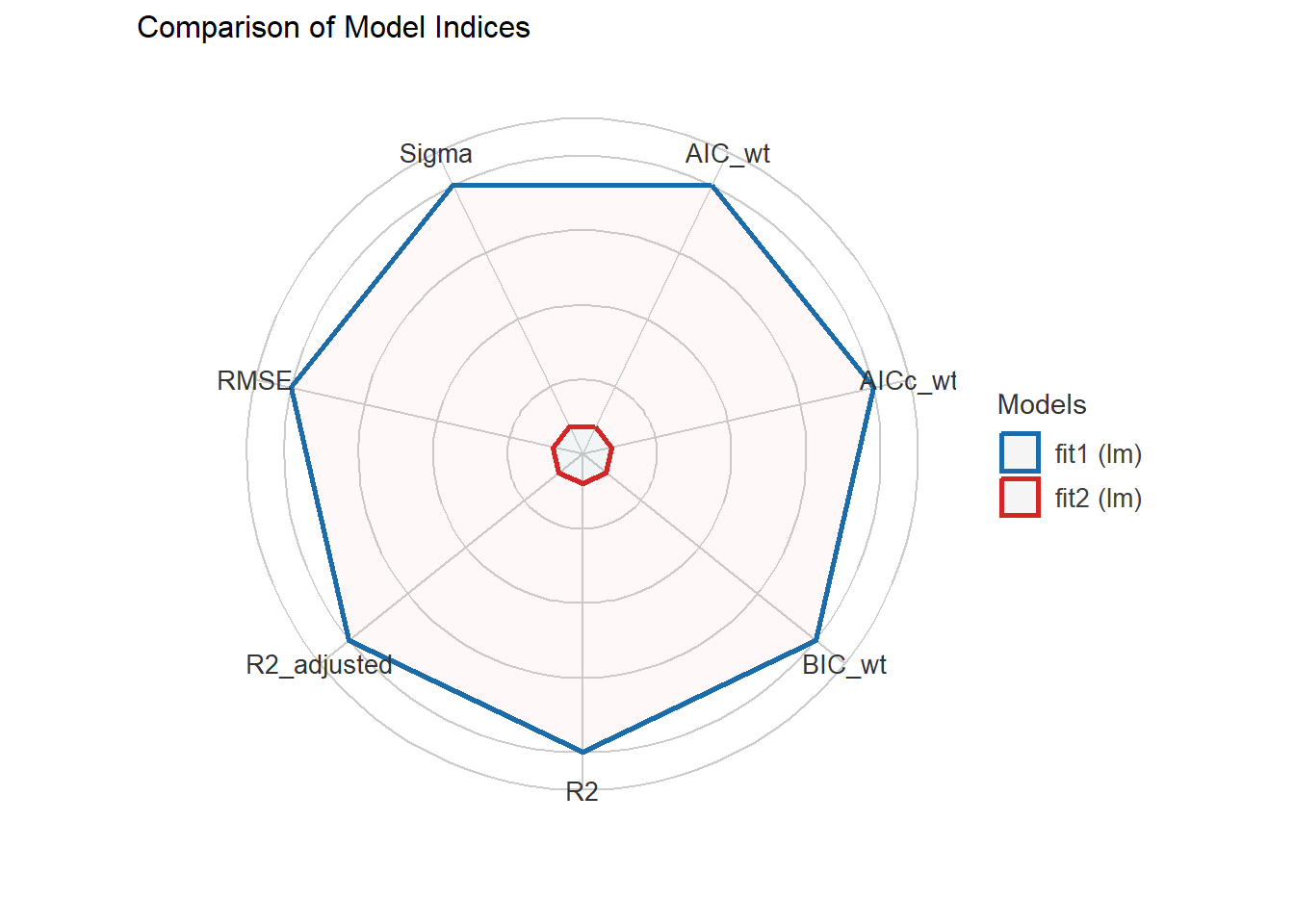
For this “spiderweb” plot, the different indices are normalized and larger values indicate better model performance. So fit2, with points closer to the center displays worse fit indices than fit1 within our training sample. Of course, this isn’t a surprise to us. For more on this, see the documentation online.
18.6 Comparing linear models (test sample)
We’ll use the augment() function from the broom package in this effort.
test1 <- augment(fit1, newdata = ccosts_test) |>
mutate(mod_n = "Model 1")
test2 <- augment(fit2, newdata = ccosts_test) |>
mutate(mod_n = "Model 2")
test_res <- bind_rows(test1, test2) |>
select(mod_n, fips, sqrt_price, .fitted, .resid,
everything()) |>
arrange(fips, mod_n)
test_res |> head()# A tibble: 6 × 11
mod_n fips sqrt_price .fitted .resid mfcc_preschool mhi_18 emp_service county
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 Mode… 10003 12.5 11.8 0.605 155. 71.0 16.5 New C…
2 Mode… 10003 12.5 11.8 0.610 155. 71.0 16.5 New C…
3 Mode… 10005 11.0 11.3 -0.270 122. 60.9 18.4 Susse…
4 Mode… 10005 11.0 11.2 -0.167 122. 60.9 18.4 Susse…
5 Mode… 1003 10.4 10.9 -0.504 109. 56.0 18.1 Baldw…
6 Mode… 1003 10.4 10.9 -0.468 109. 56.0 18.1 Baldw…
# ℹ 2 more variables: state_name <chr>, state <chr>Now, we can summarize the quality of the predictions across these two models with four summary statistics calculated across the observations in the test sample.
- the mean absolute prediction error, or MAPE
- the median absolute prediction error, or medAPE
- the maximum absolute prediction error, or maxAPE
- the square root of the mean squared prediction error, or RMSPE
No one of these dominates the other, but we might be interested in which model gives us the best (smallest) result for each of these summaries. Let’s run them.
test_res |>
group_by(mod_n) |>
summarise(MAPE = mean(abs(.resid)),
medAPE = median(abs(.resid)),
maxAPE = max(abs(.resid)),
RMSPE = sqrt(mean(.resid^2)),
rsqr = cor(sqrt_price, .fitted)^2) |>
kable()| mod_n | MAPE | medAPE | maxAPE | RMSPE | rsqr |
|---|---|---|---|---|---|
| Model 1 | 0.8054420 | 0.6643766 | 5.629447 | 1.063927 | 0.5180885 |
| Model 2 | 0.8420665 | 0.6639106 | 5.880325 | 1.126579 | 0.4540174 |
Model 1 (including both predictors) performs better within the test sample on three of these four summaries. It has the smaller mean and maximum absolute prediction error, and the smaller root mean squared prediction error. Model 2 has the smaller median absolute prediction error (though the difference is quite small.)
18.7 Bayesian linear model
Here, we’ll demonstrate a Bayesian fit including each of our predictors, with a weakly informative prior, and using the training data.
fit3 <- stan_glm(sqrt_price ~ mhi_18 + emp_service,
data = ccosts_train, refresh = 0)model_parameters(fit3, ci = 0.95)Parameter | Median | 95% CI | pd | Rhat | ESS | Prior
----------------------------------------------------------------------------------
(Intercept) | 5.32 | [4.91, 5.70] | 100% | 1.000 | 3702 | Normal (10.65 +- 3.61)
mhi_18 | 0.07 | [0.07, 0.08] | 100% | 1.000 | 4255 | Normal (0.00 +- 0.26)
emp_service | 0.09 | [0.07, 0.10] | 100% | 1.000 | 4491 | Normal (0.00 +- 1.00)model_performance(fit3)# Indices of model performance
ELPD | ELPD_SE | LOOIC | LOOIC_SE | WAIC | R2 | R2 (adj.) | RMSE | Sigma
------------------------------------------------------------------------------------
-2479.603 | 32.246 | 4959.2 | 64.493 | 4959.2 | 0.428 | 0.426 | 1.091 | 1.092check_model(fit3)